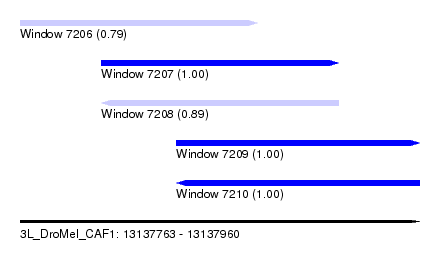
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,137,763 – 13,137,960 |
| Length | 197 |
| Max. P | 0.999262 |

| Location | 13,137,763 – 13,137,880 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.67 |
| Mean single sequence MFE | -26.98 |
| Consensus MFE | -24.31 |
| Energy contribution | -23.91 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.789330 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13137763 117 + 23771897 AUCUUUAGUCUUUGAACAAAUGUUUGUGCAUAUUUGAACUUGGCCAGGCCCAGUCUCCGAAA---AGCCCGCUCCCAAAACCAGGGUCUCCCCAGCCAAGCCAAUUUAAAAUUGGCAAAA ................(((((((......)))))))...(((((.((((((...........---((....))..........)))))).....)))))((((((.....)))))).... ( -23.60) >DroSec_CAF1 125519 117 + 1 AUCUUUAGUCUUUGAACAAAUGUUUGUGCAUAUUUGAACUUGGCCAGGCCCAGUAUCCAAAA---AGCCCGCUCCCAAAACCAGGGCCUCCCCAGCCAAGCCAAUUUAAAAUUGGCAAAA ................(((((((......)))))))...(((((.((((((...........---((....))..........)))))).....)))))((((((.....)))))).... ( -26.30) >DroSim_CAF1 125438 117 + 1 AUCUUUAGUCUUUGAACAAAUGUUUGUGCAUAUUUGAACUUGGCCAGGCCCAGUCUCCAAAA---AGCCCGCUCCCAAAACCAGGGCCUCCCCAGCCAAGCCAAUUUAAAAUUGGCAAAA ................(((((((......)))))))...(((((.((((((...........---((....))..........)))))).....)))))((((((.....)))))).... ( -26.30) >DroEre_CAF1 122288 120 + 1 AUCUUUAGUCUUUGAACAAAUGUUUGUGCAUAUUUGAACUUGGCCAGGCCCAGUCUCCAAAAAGAAGCCCCCGCCCAAAGCCGGGGUCUCCGCAGCCAGGCCAAUUUAAAAUUGGCAAAA .............((((....)))).(((...((((((.((((((.(((....(((......))).(((((.((.....)).))))).......))).))))))))))))....)))... ( -32.50) >DroYak_CAF1 126674 105 + 1 AUCUUUAGUCUUUGAACAAAUGUUUGUGCAUAUUUGAACUUGGCCAGGCCCAGUCCCCAAAACCAGG---------------AGGGUCUCCCCAGCCAAGCCAAUUUAAAAUUGGCAAAA ................(((((((......)))))))...(((((.((((((..(((.........))---------------))))))).....)))))((((((.....)))))).... ( -26.20) >consensus AUCUUUAGUCUUUGAACAAAUGUUUGUGCAUAUUUGAACUUGGCCAGGCCCAGUCUCCAAAA___AGCCCGCUCCCAAAACCAGGGUCUCCCCAGCCAAGCCAAUUUAAAAUUGGCAAAA ................(((((((......)))))))...(((((.((((((................................)))))).....)))))((((((.....)))))).... (-24.31 = -23.91 + -0.40)


| Location | 13,137,803 – 13,137,920 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.67 |
| Mean single sequence MFE | -26.60 |
| Consensus MFE | -23.97 |
| Energy contribution | -23.73 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.53 |
| SVM RNA-class probability | 0.995003 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13137803 117 + 23771897 UGGCCAGGCCCAGUCUCCGAAA---AGCCCGCUCCCAAAACCAGGGUCUCCCCAGCCAAGCCAAUUUAAAAUUGGCAAAAGGUUAAGCCAAAAAACUAACAAUCCACCGAAAAUCGAAGG ((((..(((....((...))..---.))).........((((.(((....)))......((((((.....))))))....))))..)))).............((..((.....))..)) ( -24.30) >DroSec_CAF1 125559 117 + 1 UGGCCAGGCCCAGUAUCCAAAA---AGCCCGCUCCCAAAACCAGGGCCUCCCCAGCCAAGCCAAUUUAAAAUUGGCAAAAGGUUAAGCCAAAAAACUAACAAUCCACCGAAAAUCGAAGG ((((.((((((...........---((....))..........))))))....((((..((((((.....))))))....))))..)))).............((..((.....))..)) ( -25.80) >DroSim_CAF1 125478 117 + 1 UGGCCAGGCCCAGUCUCCAAAA---AGCCCGCUCCCAAAACCAGGGCCUCCCCAGCCAAGCCAAUUUAAAAUUGGCAAAAGGUUAAGCCAAAAAACUAACAAUCCACCGAAAAUCGAAGG ((((.((((((...........---((....))..........))))))....((((..((((((.....))))))....))))..)))).............((..((.....))..)) ( -25.80) >DroEre_CAF1 122328 120 + 1 UGGCCAGGCCCAGUCUCCAAAAAGAAGCCCCCGCCCAAAGCCGGGGUCUCCGCAGCCAGGCCAAUUUAAAAUUGGCAAAAGGUUAAGCCAAAAAACUAACAAUCCACCGAAAAUCGAAGG (((((.(((....(((......))).(((((.((.....)).))))).......))).)))))........(((((..........)))))............((..((.....))..)) ( -31.40) >DroYak_CAF1 126714 105 + 1 UGGCCAGGCCCAGUCCCCAAAACCAGG---------------AGGGUCUCCCCAGCCAAGCCAAUUUAAAAUUGGCAAAAGGUUAAGCCAAAAAACUAACAAUCCACCGAAAAUCGAAGG ((((.((((((..(((.........))---------------)))))))....((((..((((((.....))))))....))))..)))).............((..((.....))..)) ( -25.70) >consensus UGGCCAGGCCCAGUCUCCAAAA___AGCCCGCUCCCAAAACCAGGGUCUCCCCAGCCAAGCCAAUUUAAAAUUGGCAAAAGGUUAAGCCAAAAAACUAACAAUCCACCGAAAAUCGAAGG ((((.((((((................................))))))....((((..((((((.....))))))....))))..)))).............((..((.....))..)) (-23.97 = -23.73 + -0.24)

| Location | 13,137,803 – 13,137,920 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.67 |
| Mean single sequence MFE | -39.22 |
| Consensus MFE | -30.10 |
| Energy contribution | -30.14 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.886104 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13137803 117 - 23771897 CCUUCGAUUUUCGGUGGAUUGUUAGUUUUUUGGCUUAACCUUUUGCCAAUUUUAAAUUGGCUUGGCUGGGGAGACCCUGGUUUUGGGAGCGGGCU---UUUCGGAGACUGGGCCUGGCCA ............((..((..(((((((....))).))))..))..))..........(((((.(((((((....))).((((((.((((......---)))).)))))).)))).))))) ( -39.40) >DroSec_CAF1 125559 117 - 1 CCUUCGAUUUUCGGUGGAUUGUUAGUUUUUUGGCUUAACCUUUUGCCAAUUUUAAAUUGGCUUGGCUGGGGAGGCCCUGGUUUUGGGAGCGGGCU---UUUUGGAUACUGGGCCUGGCCA ((.(((.....))).))...((((((((.(((((..........)))))....)))))))).(((((....((((((..((((..((((....))---))..))))...))))))))))) ( -37.80) >DroSim_CAF1 125478 117 - 1 CCUUCGAUUUUCGGUGGAUUGUUAGUUUUUUGGCUUAACCUUUUGCCAAUUUUAAAUUGGCUUGGCUGGGGAGGCCCUGGUUUUGGGAGCGGGCU---UUUUGGAGACUGGGCCUGGCCA ((.(((.....))).))...((((((((.(((((..........)))))....)))))))).(((((....((((((.((((((.((((......---)))).))))))))))))))))) ( -38.70) >DroEre_CAF1 122328 120 - 1 CCUUCGAUUUUCGGUGGAUUGUUAGUUUUUUGGCUUAACCUUUUGCCAAUUUUAAAUUGGCCUGGCUGCGGAGACCCCGGCUUUGGGCGGGGGCUUCUUUUUGGAGACUGGGCCUGGCCA ............((..((..(((((((....))).))))..))..))..........(((((.((((..((((.((((.((.....)).)))))))).....(....)..)))).))))) ( -44.50) >DroYak_CAF1 126714 105 - 1 CCUUCGAUUUUCGGUGGAUUGUUAGUUUUUUGGCUUAACCUUUUGCCAAUUUUAAAUUGGCUUGGCUGGGGAGACCCU---------------CCUGGUUUUGGGGACUGGGCCUGGCCA ((.(((.....))).))...((((((((.(((((..........)))))....)))))))).((((((((....))).---------------...((((..(....)..)))).))))) ( -35.70) >consensus CCUUCGAUUUUCGGUGGAUUGUUAGUUUUUUGGCUUAACCUUUUGCCAAUUUUAAAUUGGCUUGGCUGGGGAGACCCUGGUUUUGGGAGCGGGCU___UUUUGGAGACUGGGCCUGGCCA ............((..((..(((((((....))).))))..))..))..........(((((.(((((((....))).........................(....)..)))).))))) (-30.10 = -30.14 + 0.04)


| Location | 13,137,840 – 13,137,960 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.96 |
| Mean single sequence MFE | -33.14 |
| Consensus MFE | -30.98 |
| Energy contribution | -30.98 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.88 |
| SVM RNA-class probability | 0.997543 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13137840 120 + 23771897 CCAGGGUCUCCCCAGCCAAGCCAAUUUAAAAUUGGCAAAAGGUUAAGCCAAAAAACUAACAAUCCACCGAAAAUCGAAGGGAAAGAGAAAGGGGGAGCAGCUGGCACGCUGAUGGCAUGC .(((.(.((((((((((..((((((.....))))))....))))..................(((..((.....))...))).........)))))))..)))(((.((.....)).))) ( -33.50) >DroSec_CAF1 125596 118 + 1 CCAGGGCCUCCCCAGCCAAGCCAAUUUAAAAUUGGCAAAAGGUUAAGCCAAAAAACUAACAAUCCACCGAAAAUCGAAGGGAAAG--AAAGGGGGAGCAGCUGGCACGCUGAUGGCAUGC .(((.((..((((((((..((((((.....))))))....))))..................(((..((.....))...)))...--...))))..))..)))(((.((.....)).))) ( -34.50) >DroSim_CAF1 125515 118 + 1 CCAGGGCCUCCCCAGCCAAGCCAAUUUAAAAUUGGCAAAAGGUUAAGCCAAAAAACUAACAAUCCACCGAAAAUCGAAGGGAAAG--AAAGGGGGAGCAGCUGGCACGCUGAUGGCAUGC .(((.((..((((((((..((((((.....))))))....))))..................(((..((.....))...)))...--...))))..))..)))(((.((.....)).))) ( -34.50) >DroEre_CAF1 122368 118 + 1 CCGGGGUCUCCGCAGCCAGGCCAAUUUAAAAUUGGCAAAAGGUUAAGCCAAAAAACUAACAAUCCACCGAAAAUCGAAGGGAAGG--AAAGGGGGAGCAGCUGGCACGCUGAUGGCAUGC .(((.....))).((((..((((((.....))))))....))))..((((............(((.((.....((....))....--...)).))).((((......)))).)))).... ( -30.00) >DroYak_CAF1 126741 116 + 1 --AGGGUCUCCCCAGCCAAGCCAAUUUAAAAUUGGCAAAAGGUUAAGCCAAAAAACUAACAAUCCACCGAAAAUCGAAGGGAAGG--AAGGGGGAAGCAGCUGGCACGCUGAUGGCAUGC --...((((((((..((..((((((.....))))))....((.....)).............(((..((.....))...))).))--...)))))..((((......))))..))).... ( -33.20) >consensus CCAGGGUCUCCCCAGCCAAGCCAAUUUAAAAUUGGCAAAAGGUUAAGCCAAAAAACUAACAAUCCACCGAAAAUCGAAGGGAAAG__AAAGGGGGAGCAGCUGGCACGCUGAUGGCAUGC .(((.((..((((((((..((((((.....))))))....))))..................(((..((.....))...)))........))))..))..)))(((.((.....)).))) (-30.98 = -30.98 + 0.00)

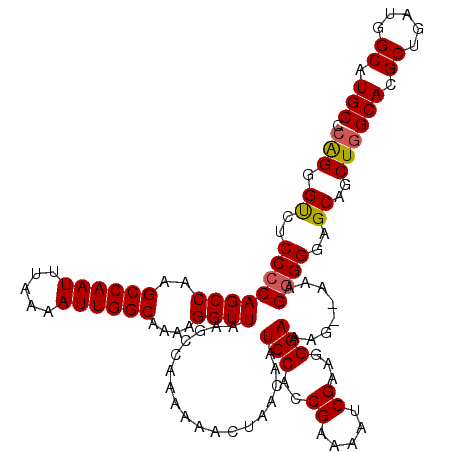
| Location | 13,137,840 – 13,137,960 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.96 |
| Mean single sequence MFE | -36.58 |
| Consensus MFE | -34.06 |
| Energy contribution | -34.50 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.93 |
| SVM decision value | 3.47 |
| SVM RNA-class probability | 0.999262 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13137840 120 - 23771897 GCAUGCCAUCAGCGUGCCAGCUGCUCCCCCUUUCUCUUUCCCUUCGAUUUUCGGUGGAUUGUUAGUUUUUUGGCUUAACCUUUUGCCAAUUUUAAAUUGGCUUGGCUGGGGAGACCCUGG ((((((.....))))))(((...((((((.........(((..(((.....))).)))..((((((((.(((((..........)))))....))))))))......))))))...))). ( -38.60) >DroSec_CAF1 125596 118 - 1 GCAUGCCAUCAGCGUGCCAGCUGCUCCCCCUUU--CUUUCCCUUCGAUUUUCGGUGGAUUGUUAGUUUUUUGGCUUAACCUUUUGCCAAUUUUAAAUUGGCUUGGCUGGGGAGGCCCUGG ((((((.....))))))(((..((..((((..(--(..(((..(((.....))).)))..((((((((.(((((..........)))))....))))))))..))..))))..)).))). ( -38.80) >DroSim_CAF1 125515 118 - 1 GCAUGCCAUCAGCGUGCCAGCUGCUCCCCCUUU--CUUUCCCUUCGAUUUUCGGUGGAUUGUUAGUUUUUUGGCUUAACCUUUUGCCAAUUUUAAAUUGGCUUGGCUGGGGAGGCCCUGG ((((((.....))))))(((..((..((((..(--(..(((..(((.....))).)))..((((((((.(((((..........)))))....))))))))..))..))))..)).))). ( -38.80) >DroEre_CAF1 122368 118 - 1 GCAUGCCAUCAGCGUGCCAGCUGCUCCCCCUUU--CCUUCCCUUCGAUUUUCGGUGGAUUGUUAGUUUUUUGGCUUAACCUUUUGCCAAUUUUAAAUUGGCCUGGCUGCGGAGACCCCGG ((((((.....))))))(((((...........--...(((..(((.....))).)))..((((((((.(((((..........)))))....))))))))..)))))(((.....))). ( -32.00) >DroYak_CAF1 126741 116 - 1 GCAUGCCAUCAGCGUGCCAGCUGCUUCCCCCUU--CCUUCCCUUCGAUUUUCGGUGGAUUGUUAGUUUUUUGGCUUAACCUUUUGCCAAUUUUAAAUUGGCUUGGCUGGGGAGACCCU-- ((((((.....))))))........((((((..--((.(((..(((.....))).)))..((((((((.(((((..........)))))....))))))))..))..)))).))....-- ( -34.70) >consensus GCAUGCCAUCAGCGUGCCAGCUGCUCCCCCUUU__CUUUCCCUUCGAUUUUCGGUGGAUUGUUAGUUUUUUGGCUUAACCUUUUGCCAAUUUUAAAUUGGCUUGGCUGGGGAGACCCUGG ((((((.....))))))(((...((((((.........(((..(((.....))).)))..((((((((.(((((..........)))))....))))))))......))))))...))). (-34.06 = -34.50 + 0.44)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:28:01 2006