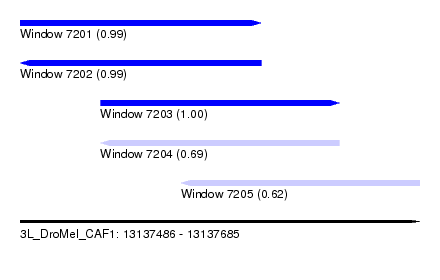
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,137,486 – 13,137,685 |
| Length | 199 |
| Max. P | 0.996190 |

| Location | 13,137,486 – 13,137,606 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.33 |
| Mean single sequence MFE | -32.26 |
| Consensus MFE | -26.84 |
| Energy contribution | -28.04 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.986890 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13137486 120 + 23771897 AUAUACCCUAGCUGCGGCCAGUCCAUCAACUGCCAGUGAUUACUGUGCUAGCAAAGACGCUACCGCAGAACGUGCAAAAAUGCAAUUGCAUAUUCAAGUUUUGCACUUGAAAUUCACCGA ...............(((.(((......)))))).((((...(((((.((((......)))).)))))...((((((((((((....)))).......)))))))).......))))... ( -32.21) >DroSec_CAF1 125240 120 + 1 AUAUUCCCUAGCUGCGGUCAGUCCAUCAACUGCCAGUGAUUACUGUGCUAGCAAAGACGCUACCGCAGAACGUGCAAAAAUUCAAUUGCAUAUUCAAGUUUUGCACUUGAAAUUCAGCGA ..........((((.((.((((......))))))........(((((.((((......)))).)))))...((((((........)))))).(((((((.....)))))))...)))).. ( -33.00) >DroSim_CAF1 125159 120 + 1 AUAUACCCUAGCUGCGGUCAGUCCAUCAACUGCCAGUGAUUACUGUGCUAGCAAAGACGCUACCGCAGAACGUGCAAAAAUGCAAUUGCAUAUUCAAGUUUUGCACUUGAAAUUCAGCGA ..........((((.((.((((......)))))).(..(((.(((((.((((......)))).))))).....(((....))))))..)...(((((((.....)))))))...)))).. ( -33.90) >DroEre_CAF1 122012 120 + 1 AUAUGCCCCAGCUGCGGUCAGUCCAUCAACUGCCAGUGAGUACUGUGCUAGCAAAGACGCUACCGCAGAACGUGCAAAAAUGCAAUUGCAUAUUCAAGUUUUGCACUUGAAAUUCACCGA ...............((.((((......)))))).((((((.(((((.((((......)))).)))))...((((((((((((....)))).......)))))))).....))))))... ( -34.41) >DroYak_CAF1 126404 111 + 1 AUAUGCCCCAGCUGCGGUCAGUCCAUCAACUGCCAGUGAUUA---------CAAAGACGCUACCGCAGAACGUGCAAAAAUGCAAUUGCAUAUUCAAGUUUUGCACUUGAAAUUCACCGA .(((((.....(((((((((((......))))..((((....---------......)))))))))))....((((....))))...)))))(((((((.....)))))))......... ( -27.80) >consensus AUAUACCCUAGCUGCGGUCAGUCCAUCAACUGCCAGUGAUUACUGUGCUAGCAAAGACGCUACCGCAGAACGUGCAAAAAUGCAAUUGCAUAUUCAAGUUUUGCACUUGAAAUUCACCGA ...........(((((((..(((........((((((....)))).)).......)))...)))))))...((((((........)))))).(((((((.....)))))))......... (-26.84 = -28.04 + 1.20)


| Location | 13,137,486 – 13,137,606 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.33 |
| Mean single sequence MFE | -37.88 |
| Consensus MFE | -32.16 |
| Energy contribution | -33.40 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.991126 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13137486 120 - 23771897 UCGGUGAAUUUCAAGUGCAAAACUUGAAUAUGCAAUUGCAUUUUUGCACGUUCUGCGGUAGCGUCUUUGCUAGCACAGUAAUCACUGGCAGUUGAUGGACUGGCCGCAGCUAGGGUAUAU .........(((((((.....)))))))(((((...((((....))))(...(((((((...((((..(((.((.((((....)))))))))....))))..)))))))...).))))). ( -38.80) >DroSec_CAF1 125240 120 - 1 UCGCUGAAUUUCAAGUGCAAAACUUGAAUAUGCAAUUGAAUUUUUGCACGUUCUGCGGUAGCGUCUUUGCUAGCACAGUAAUCACUGGCAGUUGAUGGACUGACCGCAGCUAGGGAAUAU ((.(((........((((((((.((.(((.....))).)).))))))))...(((((((...((((..(((.((.((((....)))))))))....))))..))))))).))).)).... ( -38.90) >DroSim_CAF1 125159 120 - 1 UCGCUGAAUUUCAAGUGCAAAACUUGAAUAUGCAAUUGCAUUUUUGCACGUUCUGCGGUAGCGUCUUUGCUAGCACAGUAAUCACUGGCAGUUGAUGGACUGACCGCAGCUAGGGUAUAU .........(((((((.....)))))))(((((...((((....))))(...(((((((...((((..(((.((.((((....)))))))))....))))..)))))))...).))))). ( -38.50) >DroEre_CAF1 122012 120 - 1 UCGGUGAAUUUCAAGUGCAAAACUUGAAUAUGCAAUUGCAUUUUUGCACGUUCUGCGGUAGCGUCUUUGCUAGCACAGUACUCACUGGCAGUUGAUGGACUGACCGCAGCUGGGGCAUAU (((((((.......((((((((.......((((....))))))))))))((.(((.(.(((((....))))).).))).)))))))))((((((.(((.....)))))))))........ ( -40.81) >DroYak_CAF1 126404 111 - 1 UCGGUGAAUUUCAAGUGCAAAACUUGAAUAUGCAAUUGCAUUUUUGCACGUUCUGCGGUAGCGUCUUUG---------UAAUCACUGGCAGUUGAUGGACUGACCGCAGCUGGGGCAUAU (((((((..(((((((.....)))))))..(((((.(((....(((((.....)))))..)))...)))---------)).)))))))((((((.(((.....)))))))))........ ( -32.40) >consensus UCGGUGAAUUUCAAGUGCAAAACUUGAAUAUGCAAUUGCAUUUUUGCACGUUCUGCGGUAGCGUCUUUGCUAGCACAGUAAUCACUGGCAGUUGAUGGACUGACCGCAGCUAGGGCAUAU .........(((((((.....)))))))(((((...((((....))))(.(.(((((((...((((..(((.((.((((....)))))))))....))))..)))))))..).)))))). (-32.16 = -33.40 + 1.24)


| Location | 13,137,526 – 13,137,645 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.32 |
| Mean single sequence MFE | -31.78 |
| Consensus MFE | -27.82 |
| Energy contribution | -29.62 |
| Covariance contribution | 1.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.66 |
| SVM RNA-class probability | 0.996190 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13137526 119 + 23771897 UACUGUGCUAGCAAAGACGCUACCGCAGAACGUGCAAAAAUGCAAUUGCAUAUUCAAGUUUUGCACUUGAAAUUCACCGAUUUGUAUCGUUCGGCGCUCGUAUGAAUGCCCAAA-AAAGC ...((((.((((......)))).))))(((((((((((.((((....)))).(((((((.....))))))).........)))))).)))))((((.((....)).))))....-..... ( -34.20) >DroSec_CAF1 125280 120 + 1 UACUGUGCUAGCAAAGACGCUACCGCAGAACGUGCAAAAAUUCAAUUGCAUAUUCAAGUUUUGCACUUGAAAUUCAGCGAUUUGUAUCGUUCGGCGCUCGUAUGAAUGCCCAAAAAAAGC ..(((((.((((......)))).)))))...((((((........)))))).(((((((.....)))))))....((((((....)))))).((((.((....)).)))).......... ( -31.80) >DroSim_CAF1 125199 120 + 1 UACUGUGCUAGCAAAGACGCUACCGCAGAACGUGCAAAAAUGCAAUUGCAUAUUCAAGUUUUGCACUUGAAAUUCAGCGAUUUGUAUCGUUCGGCGCUCGUAUGAAUGCCCAAAAAAAGC ...((((.((((......)))).))))(((((((((((.((((....)))).(((((((.....))))))).........)))))).)))))((((.((....)).)))).......... ( -34.20) >DroEre_CAF1 122052 119 + 1 UACUGUGCUAGCAAAGACGCUACCGCAGAACGUGCAAAAAUGCAAUUGCAUAUUCAAGUUUUGCACUUGAAAUUCACCGAUUUGUAUCGCUCGGCGCUCGUAUGAAUGCCCAAA-AAAGC ..(((((.((((......)))).)))))..((((((((.((((....)))).(((((((.....))))))).........)))))).))...((((.((....)).))))....-..... ( -31.00) >DroYak_CAF1 126444 110 + 1 UA---------CAAAGACGCUACCGCAGAACGUGCAAAAAUGCAAUUGCAUAUUCAAGUUUUGCACUUGAAAUUCACCGAUUUGUAUCGUUCGGCGCUCGUAUGAAUGCCCAAA-AAAGC ..---------.......(((......(((((((((((.((((....)))).(((((((.....))))))).........)))))).)))))((((.((....)).))))....-..))) ( -27.70) >consensus UACUGUGCUAGCAAAGACGCUACCGCAGAACGUGCAAAAAUGCAAUUGCAUAUUCAAGUUUUGCACUUGAAAUUCACCGAUUUGUAUCGUUCGGCGCUCGUAUGAAUGCCCAAA_AAAGC ...((((.((((......)))).))))(((((((((((.((((....)))).(((((((.....))))))).........)))))).)))))((((.((....)).)))).......... (-27.82 = -29.62 + 1.80)


| Location | 13,137,526 – 13,137,645 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.32 |
| Mean single sequence MFE | -31.44 |
| Consensus MFE | -28.96 |
| Energy contribution | -29.60 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.688592 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13137526 119 - 23771897 GCUUU-UUUGGGCAUUCAUACGAGCGCCGAACGAUACAAAUCGGUGAAUUUCAAGUGCAAAACUUGAAUAUGCAAUUGCAUUUUUGCACGUUCUGCGGUAGCGUCUUUGCUAGCACAGUA ((((.-...)))).....((((((((((((..........))))).........((((((((.......((((....))))))))))))))))((.(.(((((....))))).).))))) ( -32.01) >DroSec_CAF1 125280 120 - 1 GCUUUUUUUGGGCAUUCAUACGAGCGCCGAACGAUACAAAUCGCUGAAUUUCAAGUGCAAAACUUGAAUAUGCAAUUGAAUUUUUGCACGUUCUGCGGUAGCGUCUUUGCUAGCACAGUA ((........(((.(((....))).)))(((((................(((((((.....)))))))..(((((........)))))))))).))(.(((((....))))).)...... ( -32.00) >DroSim_CAF1 125199 120 - 1 GCUUUUUUUGGGCAUUCAUACGAGCGCCGAACGAUACAAAUCGCUGAAUUUCAAGUGCAAAACUUGAAUAUGCAAUUGCAUUUUUGCACGUUCUGCGGUAGCGUCUUUGCUAGCACAGUA ((........(((.(((....))).)))(((((.........((.....(((((((.....)))))))...))...((((....))))))))).))(.(((((....))))).)...... ( -33.50) >DroEre_CAF1 122052 119 - 1 GCUUU-UUUGGGCAUUCAUACGAGCGCCGAGCGAUACAAAUCGGUGAAUUUCAAGUGCAAAACUUGAAUAUGCAAUUGCAUUUUUGCACGUUCUGCGGUAGCGUCUUUGCUAGCACAGUA ((((.-...)))).....((((((((((((..........))))).........((((((((.......((((....))))))))))))))))((.(.(((((....))))).).))))) ( -32.81) >DroYak_CAF1 126444 110 - 1 GCUUU-UUUGGGCAUUCAUACGAGCGCCGAACGAUACAAAUCGGUGAAUUUCAAGUGCAAAACUUGAAUAUGCAAUUGCAUUUUUGCACGUUCUGCGGUAGCGUCUUUG---------UA (((..-.((((((.(((....))).)))(((((................(((((((.....)))))))..(((((........))))))))))..))).))).......---------.. ( -26.90) >consensus GCUUU_UUUGGGCAUUCAUACGAGCGCCGAACGAUACAAAUCGGUGAAUUUCAAGUGCAAAACUUGAAUAUGCAAUUGCAUUUUUGCACGUUCUGCGGUAGCGUCUUUGCUAGCACAGUA ((........(((.(((....))).)))(((((................(((((((.....)))))))..(((((........)))))))))).))(.(((((....))))).)...... (-28.96 = -29.60 + 0.64)


| Location | 13,137,566 – 13,137,685 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.41 |
| Mean single sequence MFE | -34.99 |
| Consensus MFE | -30.04 |
| Energy contribution | -30.72 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.618042 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13137566 119 - 23771897 GUUUCGUGCCCAGGUUUUCCCAUUUUCCAGUGGGGGAGUAGCUUU-UUUGGGCAUUCAUACGAGCGCCGAACGAUACAAAUCGGUGAAUUUCAAGUGCAAAACUUGAAUAUGCAAUUGCA ((...(((((((((((..(((((......)))))..)).......-)))))))))....))...((((((..........))))))...(((((((.....)))))))..(((....))) ( -36.31) >DroSec_CAF1 125320 120 - 1 GUUUCGUGCCCAGGUUUUCCCAUUUUCCAGUGGGGGAGUAGCUUUUUUUGGGCAUUCAUACGAGCGCCGAACGAUACAAAUCGCUGAAUUUCAAGUGCAAAACUUGAAUAUGCAAUUGAA ((.(((((((((((((..(((((......)))))..))........))))))).......((.....)).)))).)).....((.....(((((((.....)))))))...))....... ( -33.00) >DroSim_CAF1 125239 120 - 1 GUUUCGUGCCCAGGUUUUCCCAUUUUCCAGUGGGGGAGUAGCUUUUUUUGGGCAUUCAUACGAGCGCCGAACGAUACAAAUCGCUGAAUUUCAAGUGCAAAACUUGAAUAUGCAAUUGCA ((.(((((((((((((..(((((......)))))..))........))))))).......((.....)).)))).)).....((.....(((((((.....)))))))...))....... ( -33.00) >DroEre_CAF1 122092 119 - 1 GUUUCGUGCCCAGGUUUUCCUAUUUUCCAGCGGGGGAGUAGCUUU-UUUGGGCAUUCAUACGAGCGCCGAGCGAUACAAAUCGGUGAAUUUCAAGUGCAAAACUUGAAUAUGCAAUUGCA ((...(((((((((.....(((((..((....))..)))))....-)))))))))....))...((((((..........))))))...(((((((.....)))))))..(((....))) ( -38.40) >DroYak_CAF1 126475 119 - 1 GUUUCGUGCCCAGGUUUUCCGAUUUUCCAGUGGGGGAGUAGCUUU-UUUGGGCAUUCAUACGAGCGCCGAACGAUACAAAUCGGUGAAUUUCAAGUGCAAAACUUGAAUAUGCAAUUGCA ((...(((((((((.......(((..((....))..)))......-)))))))))....))...((((((..........))))))...(((((((.....)))))))..(((....))) ( -34.22) >consensus GUUUCGUGCCCAGGUUUUCCCAUUUUCCAGUGGGGGAGUAGCUUU_UUUGGGCAUUCAUACGAGCGCCGAACGAUACAAAUCGGUGAAUUUCAAGUGCAAAACUUGAAUAUGCAAUUGCA ((...(((((((((((..(((((......)))))..))........)))))))))....))..(((((((..........)))))....(((((((.....)))))))...))....... (-30.04 = -30.72 + 0.68)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:27:56 2006