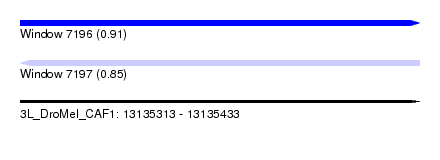
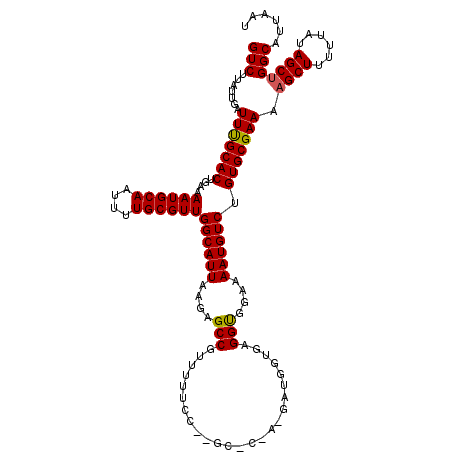
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,135,313 – 13,135,433 |
| Length | 120 |
| Max. P | 0.908169 |

| Location | 13,135,313 – 13,135,433 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.85 |
| Mean single sequence MFE | -25.91 |
| Consensus MFE | -19.51 |
| Energy contribution | -19.51 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908169 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13135313 120 + 23771897 AUUAAUGCCAGCUAUGAAAAGCUUUUCGCACAGACAUUUUCCACCUCACCAUCGUCGUGCCGGGAAAAACGGCUCUUAAUGCCAACGCAAAAUUGCAUUUUCAGUGCAAAUCAAUAAGAC ......(((.(((......)))((..(((((.(((..................)))))).))..))....)))(((((.(((....)))...((((((.....)))))).....))))). ( -25.67) >DroSec_CAF1 123107 107 + 1 AUUAAUGCCAGCUAUAAAAAGCUUUUCGCACGGACAUUUUCCACCUGAC-------------GGAAAAACGGCUCUUAAUGCCAACGCAAAAUUGCAUUUUCAGUGCGAAUCAAUAAGAC .........((((......)))).((((((((((.....))).......-------------.(((((..(((.......)))...(((....))).))))).))))))).......... ( -25.70) >DroSim_CAF1 122986 107 + 1 AUUAAUGCCAGCUAUAAAAAGCUUUUCGCACAGACAUUUUCCACCUGAC-------------GGAAAAACGGCUCUUAAUGCCAACGCAAAAUUGCAUUUUCAGUGCGAAUCAAUAAGAC .........((((......)))).(((((((.....((((((.......-------------))))))..(((.......)))...(((....))).......))))))).......... ( -24.80) >DroEre_CAF1 120014 118 + 1 AUUAAUGCCAGCUAUAAAAAGCUUUUCGCACAGACAUUUUCCGCCUCACCAUCCUCGGGCU-GGAA-AACGGCUCUUAAUGCCAACGCAAAAUUGCAUUUUCAGUGCAAAUCAAUAAGAC ......(((((((......)))).............((((((((((..........)))).-))))-)).)))(((((.(((....)))...((((((.....)))))).....))))). ( -27.30) >DroYak_CAF1 124187 119 + 1 AUUAAUGCCGGCUAUAAAAAGCUUUUCGCACAGACAUUCUCCACCUCACCAUCCUUGGGCU-GGAAAAACGGCUCUUAAUGCCAACGCAAAAUUGCAUUUUCAGUGCAAAUCAAUAAGAC ......(((((((......))).................((((.((((.......)))).)-)))....))))(((((.(((....)))...((((((.....)))))).....))))). ( -26.10) >consensus AUUAAUGCCAGCUAUAAAAAGCUUUUCGCACAGACAUUUUCCACCUCACCAUC_U_G_GC__GGAAAAACGGCUCUUAAUGCCAACGCAAAAUUGCAUUUUCAGUGCAAAUCAAUAAGAC ......(((((((......)))).............((((((....................))))))..)))(((((.(((....)))...((((((.....)))))).....))))). (-19.51 = -19.51 + -0.00)

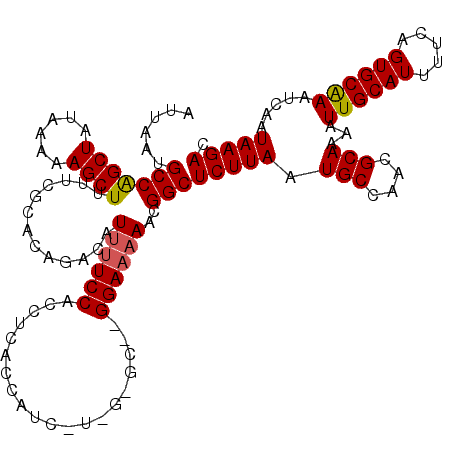
| Location | 13,135,313 – 13,135,433 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.85 |
| Mean single sequence MFE | -32.46 |
| Consensus MFE | -26.91 |
| Energy contribution | -26.71 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.853357 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13135313 120 - 23771897 GUCUUAUUGAUUUGCACUGAAAAUGCAAUUUUGCGUUGGCAUUAAGAGCCGUUUUUCCCGGCACGACGAUGGUGAGGUGGAAAAUGUCUGUGCGAAAAGCUUUUCAUAGCUGGCAUUAAU (((.......(((((((((..((((((....))))))..)).....((.((((((((((..(((.......))).)).)))))))).))))))))).((((......)))))))...... ( -33.80) >DroSec_CAF1 123107 107 - 1 GUCUUAUUGAUUCGCACUGAAAAUGCAAUUUUGCGUUGGCAUUAAGAGCCGUUUUUCC-------------GUCAGGUGGAAAAUGUCCGUGCGAAAAGCUUUUUAUAGCUGGCAUUAAU (((.......(((((((.((.((((((....))))))(((.......)))..((((((-------------(.....)))))))..)).))))))).((((......)))))))...... ( -30.60) >DroSim_CAF1 122986 107 - 1 GUCUUAUUGAUUCGCACUGAAAAUGCAAUUUUGCGUUGGCAUUAAGAGCCGUUUUUCC-------------GUCAGGUGGAAAAUGUCUGUGCGAAAAGCUUUUUAUAGCUGGCAUUAAU (((.......(((((((.((.((((((....))))))(((.......)))..((((((-------------(.....)))))))..)).))))))).((((......)))))))...... ( -30.60) >DroEre_CAF1 120014 118 - 1 GUCUUAUUGAUUUGCACUGAAAAUGCAAUUUUGCGUUGGCAUUAAGAGCCGUU-UUCC-AGCCCGAGGAUGGUGAGGCGGAAAAUGUCUGUGCGAAAAGCUUUUUAUAGCUGGCAUUAAU (((.......(((((((....((((((....))))))((((((.....(((((-((((-(.((...)).))).)))))))..)))))).))))))).((((......)))))))...... ( -34.90) >DroYak_CAF1 124187 119 - 1 GUCUUAUUGAUUUGCACUGAAAAUGCAAUUUUGCGUUGGCAUUAAGAGCCGUUUUUCC-AGCCCAAGGAUGGUGAGGUGGAGAAUGUCUGUGCGAAAAGCUUUUUAUAGCCGGCAUUAAU (((.......(((((((....((((((....))))))((((((.....(((..((.((-(.((...)).))).))..)))..)))))).)))))))..(((......))).)))...... ( -32.40) >consensus GUCUUAUUGAUUUGCACUGAAAAUGCAAUUUUGCGUUGGCAUUAAGAGCCGUUUUUCC__GC_C_A_GAUGGUGAGGUGGAAAAUGUCUGUGCGAAAAGCUUUUUAUAGCUGGCAUUAAU (((.......(((((((....((((((....))))))((((((....(((.........................)))....)))))).))))))).((((......)))))))...... (-26.91 = -26.71 + -0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:27:48 2006