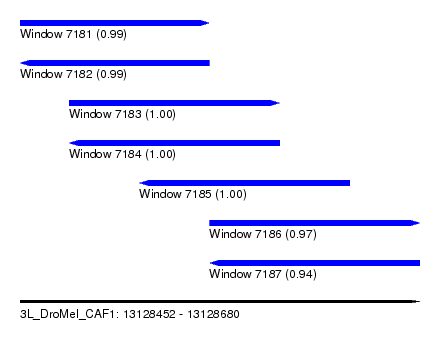
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,128,452 – 13,128,680 |
| Length | 228 |
| Max. P | 0.999873 |

| Location | 13,128,452 – 13,128,560 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 96.45 |
| Mean single sequence MFE | -36.65 |
| Consensus MFE | -36.59 |
| Energy contribution | -36.15 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.72 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.19 |
| SVM RNA-class probability | 0.990054 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13128452 108 + 23771897 GUUUUUUGUUAUUAUUGUUGGCUGGCUGCAAGGUUGGUGGUGAGGCAAGUGUAUUUGUUUAUUGGCCUCAUUGCCGGGGUUACAGCGAAUUGCAUUUGCCACGGGCAA .....(((((.((((..(..(((........)))..)..))))((((((((((((((((...(((((((......))))))).)))))).))))))))))...))))) ( -37.30) >DroSec_CAF1 116290 108 + 1 GGUUUUUGUUGUUAUUGUCGGGUGGCUGCAAGGUUGGUGGUGAGGCAAGUGUAUUUGUUUAUUGGCCUCAUUGCCGGGGUUACAGCGAAUUGCAUUUGCCACGGGCAA ..............(((((..((((((((((..((((..(((((((.((((........)))).)))))))..)))).((....))...)))))...))))).))))) ( -37.30) >DroSim_CAF1 114567 108 + 1 GUUUUUUGUUAUUAUUGUCGGGUGGCUGCAAGGUUGGUGGUGAGGCAAGUGUAUUUGUUUAUUGGCCUCAUUGCCGGGGUUACAGCGAAUUGCAUUUGCCACGGGCAA ..............(((((..((((((((((..((((..(((((((.((((........)))).)))))))..)))).((....))...)))))...))))).))))) ( -37.30) >DroEre_CAF1 113297 107 + 1 GUUUUUUGU-GUUGUUGUUGGGUGGCUGCAAGGUUGGUGGUGGGGCAAGUGUAUUUGUUUAUUGGCCUCAUUGCCGGGGUUACAGCGAAUUGCAUUUGCCACGGGCAA .........-....(((((.(.(((((((((..((((..(((((((.((((........)))).)))))))..)))).((....))...)))))...))))).))))) ( -34.70) >consensus GUUUUUUGUUAUUAUUGUCGGGUGGCUGCAAGGUUGGUGGUGAGGCAAGUGUAUUUGUUUAUUGGCCUCAUUGCCGGGGUUACAGCGAAUUGCAUUUGCCACGGGCAA ..............(((((.(.(((((((((..((((..(((((((.((((........)))).)))))))..)))).((....))...)))))...))))).))))) (-36.59 = -36.15 + -0.44)

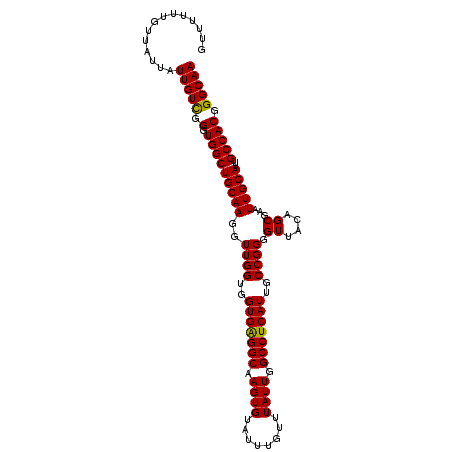
| Location | 13,128,452 – 13,128,560 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 96.45 |
| Mean single sequence MFE | -22.72 |
| Consensus MFE | -21.69 |
| Energy contribution | -22.20 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.35 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.987574 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13128452 108 - 23771897 UUGCCCGUGGCAAAUGCAAUUCGCUGUAACCCCGGCAAUGAGGCCAAUAAACAAAUACACUUGCCUCACCACCAACCUUGCAGCCAGCCAACAAUAAUAACAAAAAAC .......((((...(((((...((((......))))..((((((..................)))))).........)))))....)))).................. ( -22.87) >DroSec_CAF1 116290 108 - 1 UUGCCCGUGGCAAAUGCAAUUCGCUGUAACCCCGGCAAUGAGGCCAAUAAACAAAUACACUUGCCUCACCACCAACCUUGCAGCCACCCGACAAUAACAACAAAAACC ......(((((...(((((...((((......))))..((((((..................)))))).........))))))))))..................... ( -24.27) >DroSim_CAF1 114567 108 - 1 UUGCCCGUGGCAAAUGCAAUUCGCUGUAACCCCGGCAAUGAGGCCAAUAAACAAAUACACUUGCCUCACCACCAACCUUGCAGCCACCCGACAAUAAUAACAAAAAAC ......(((((...(((((...((((......))))..((((((..................)))))).........))))))))))..................... ( -24.27) >DroEre_CAF1 113297 107 - 1 UUGCCCGUGGCAAAUGCAAUUCGCUGUAACCCCGGCAAUGAGGCCAAUAAACAAAUACACUUGCCCCACCACCAACCUUGCAGCCACCCAACAACAAC-ACAAAAAAC ......(((((...(((((...((((......))))..((.(((..................))).)).........))))))))))...........-......... ( -19.47) >consensus UUGCCCGUGGCAAAUGCAAUUCGCUGUAACCCCGGCAAUGAGGCCAAUAAACAAAUACACUUGCCUCACCACCAACCUUGCAGCCACCCAACAAUAACAACAAAAAAC ......(((((...(((((...((((......))))..((((((..................)))))).........))))))))))..................... (-21.69 = -22.20 + 0.50)

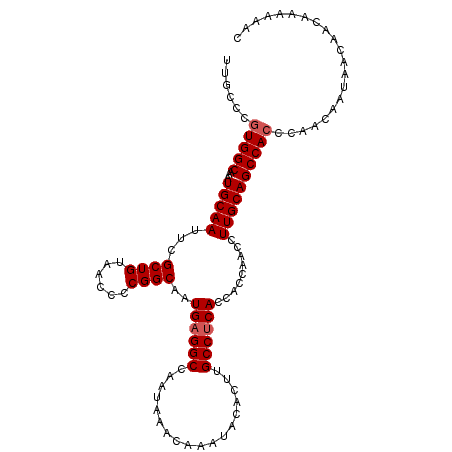
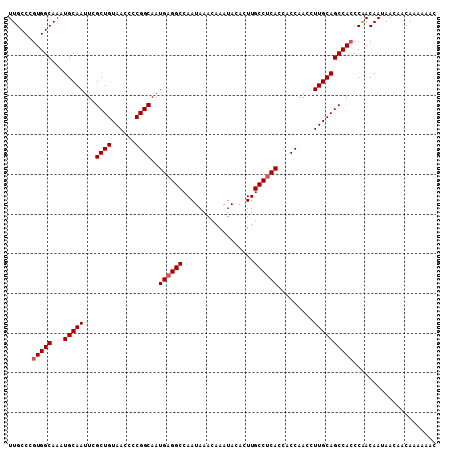
| Location | 13,128,480 – 13,128,600 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.00 |
| Mean single sequence MFE | -48.74 |
| Consensus MFE | -46.16 |
| Energy contribution | -46.20 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.02 |
| Mean z-score | -3.18 |
| Structure conservation index | 0.95 |
| SVM decision value | 4.33 |
| SVM RNA-class probability | 0.999873 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13128480 120 + 23771897 CAAGGUUGGUGGUGAGGCAAGUGUAUUUGUUUAUUGGCCUCAUUGCCGGGGUUACAGCGAAUUGCAUUUGCCACGGGCAACUAAACCCUCGACAUCCUUAACAGGAUGUCCCGCCACUUG ((((..((((((.((((((((((((((((((...(((((((......))))))).)))))).))))))))))..(((........)))))((((((((....)))))))))))))))))) ( -51.50) >DroSec_CAF1 116318 120 + 1 CAAGGUUGGUGGUGAGGCAAGUGUAUUUGUUUAUUGGCCUCAUUGCCGGGGUUACAGCGAAUUGCAUUUGCCACGGGCAACUAAACCCUCGACAUCCUUGACAGGAUGCCCCGCCACUUG ((((..(((((((((((((((((((((((((...(((((((......))))))).)))))).))))))))))..(((........)))))).((((((....))))))..)))))))))) ( -46.90) >DroSim_CAF1 114595 120 + 1 CAAGGUUGGUGGUGAGGCAAGUGUAUUUGUUUAUUGGCCUCAUUGCCGGGGUUACAGCGAAUUGCAUUUGCCACGGGCAACUAAACCCUCGACAUCCUUGACAGGAUGCCCCGCCACUUG ((((..(((((((((((((((((((((((((...(((((((......))))))).)))))).))))))))))..(((........)))))).((((((....))))))..)))))))))) ( -46.90) >DroEre_CAF1 113324 119 + 1 CAAGGUUGGUGGUGGGGCAAGUGUAUUUGUUUAUUGGCCUCAUUGCCGGGGUUACAGCGAAUUGCAUUUGCCACGGGCAACUAAACCCUCGACAUCCUCGACAGGAUGCCCCGCCACU-G .......((((((((((((((((((((((((...(((((((......))))))).)))))).))))))))))..(((........)))....((((((....))))))..))))))))-. ( -49.80) >DroYak_CAF1 116777 120 + 1 CAAGGUGGGUGGUGAGGCAAGUGUAUUUGUUUAUUGGCCUCAUUGCCGGGGUUACAGCGAAUUGCAUUUGCCACGGGCAACUAAACCCUCGACAUCCUUGACAGGAUGCCCCGCCACUUG ...((((((.(((..((((((((((((((((...(((((((......))))))).)))))).))))))))))..((....))..))).....((((((....)))))).))))))..... ( -48.60) >consensus CAAGGUUGGUGGUGAGGCAAGUGUAUUUGUUUAUUGGCCUCAUUGCCGGGGUUACAGCGAAUUGCAUUUGCCACGGGCAACUAAACCCUCGACAUCCUUGACAGGAUGCCCCGCCACUUG ..((((.((((((((((((((((((((((((...(((((((......))))))).)))))).))))))))))..(((........)))))).((((((....))))))..))))))))). (-46.16 = -46.20 + 0.04)

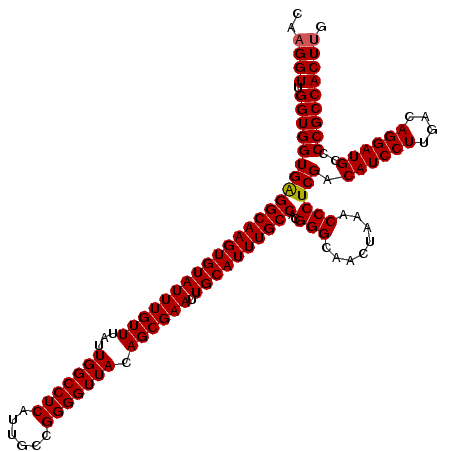

| Location | 13,128,480 – 13,128,600 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.00 |
| Mean single sequence MFE | -41.21 |
| Consensus MFE | -38.96 |
| Energy contribution | -39.20 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.84 |
| SVM RNA-class probability | 0.997332 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13128480 120 - 23771897 CAAGUGGCGGGACAUCCUGUUAAGGAUGUCGAGGGUUUAGUUGCCCGUGGCAAAUGCAAUUCGCUGUAACCCCGGCAAUGAGGCCAAUAAACAAAUACACUUGCCUCACCACCAACCUUG (((((((.((((((((((....))))))))((((((((.((((((.(.((.....((.....)).....))).)))))).)))))......(((......))).))).)).)))..)))) ( -41.60) >DroSec_CAF1 116318 120 - 1 CAAGUGGCGGGGCAUCCUGUCAAGGAUGUCGAGGGUUUAGUUGCCCGUGGCAAAUGCAAUUCGCUGUAACCCCGGCAAUGAGGCCAAUAAACAAAUACACUUGCCUCACCACCAACCUUG (((((((.((((((((((....))))))))((((((((.((((((.(.((.....((.....)).....))).)))))).)))))......(((......))).))).)).)))..)))) ( -41.00) >DroSim_CAF1 114595 120 - 1 CAAGUGGCGGGGCAUCCUGUCAAGGAUGUCGAGGGUUUAGUUGCCCGUGGCAAAUGCAAUUCGCUGUAACCCCGGCAAUGAGGCCAAUAAACAAAUACACUUGCCUCACCACCAACCUUG (((((((.((((((((((....))))))))((((((((.((((((.(.((.....((.....)).....))).)))))).)))))......(((......))).))).)).)))..)))) ( -41.00) >DroEre_CAF1 113324 119 - 1 C-AGUGGCGGGGCAUCCUGUCGAGGAUGUCGAGGGUUUAGUUGCCCGUGGCAAAUGCAAUUCGCUGUAACCCCGGCAAUGAGGCCAAUAAACAAAUACACUUGCCCCACCACCAACCUUG .-.((((.((((((((((....))))(((((.(((((((((((((...))))..........)))).))))))))))........................)))))).))))........ ( -42.00) >DroYak_CAF1 116777 120 - 1 CAAGUGGCGGGGCAUCCUGUCAAGGAUGUCGAGGGUUUAGUUGCCCGUGGCAAAUGCAAUUCGCUGUAACCCCGGCAAUGAGGCCAAUAAACAAAUACACUUGCCUCACCACCCACCUUG .....((.((((((((((....))))))))..((((......))))((((............((((......))))..((((((..................)))))))))))).))... ( -40.47) >consensus CAAGUGGCGGGGCAUCCUGUCAAGGAUGUCGAGGGUUUAGUUGCCCGUGGCAAAUGCAAUUCGCUGUAACCCCGGCAAUGAGGCCAAUAAACAAAUACACUUGCCUCACCACCAACCUUG (((((((.((((((((((....))))))))..((((......))))(.(((((.((((......)))).....(((......)))...............))))).).)).)))..)))) (-38.96 = -39.20 + 0.24)

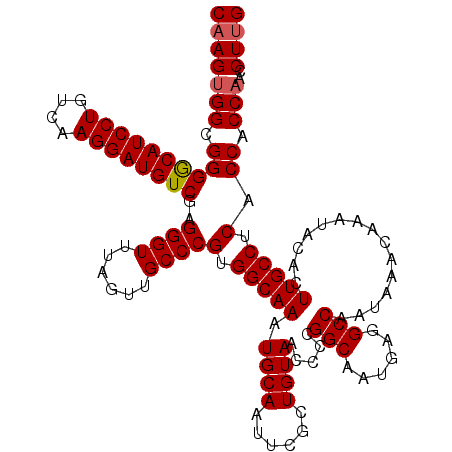
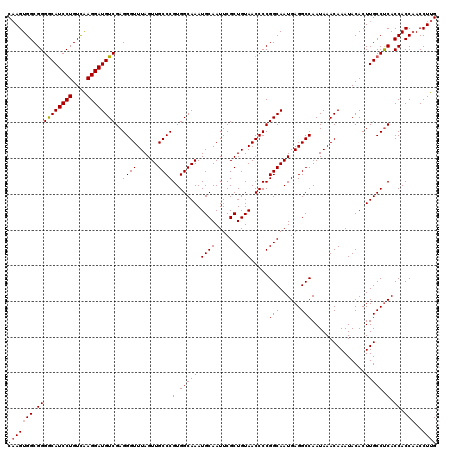
| Location | 13,128,520 – 13,128,640 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.67 |
| Mean single sequence MFE | -41.88 |
| Consensus MFE | -39.96 |
| Energy contribution | -40.04 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.95 |
| SVM decision value | 3.23 |
| SVM RNA-class probability | 0.998786 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13128520 120 - 23771897 AUAAACCCAUAAAGUUUAACUCGCAUUAGUUGAAGGCAGCCAAGUGGCGGGACAUCCUGUUAAGGAUGUCGAGGGUUUAGUUGCCCGUGGCAAAUGCAAUUCGCUGUAACCCCGGCAAUG .(((((((......(((((((......)))))))....(((....)))..((((((((....))))))))..)))))))((((((.(.((.....((.....)).....))).)))))). ( -42.30) >DroSec_CAF1 116358 120 - 1 AUAAACCCAUAACGUUUAACUCGCAUUAGUUGAAGGCAGCCAAGUGGCGGGGCAUCCUGUCAAGGAUGUCGAGGGUUUAGUUGCCCGUGGCAAAUGCAAUUCGCUGUAACCCCGGCAAUG .(((((((....(.(((((((......))))))).)..(((....)))..((((((((....))))))))..)))))))((((((.(.((.....((.....)).....))).)))))). ( -43.10) >DroSim_CAF1 114635 120 - 1 AUAAACGCAUAAAGUUUAACUCGCAUUAGUUGAAGGCAGCCAAGUGGCGGGGCAUCCUGUCAAGGAUGUCGAGGGUUUAGUUGCCCGUGGCAAAUGCAAUUCGCUGUAACCCCGGCAAUG .(((((.......)))))....(((((.((((..(((((((....)))..((((((((....))))))))...........))))..)))).))))).....((((......)))).... ( -39.10) >DroEre_CAF1 113364 119 - 1 AUAAACCCAUAAAGUUUAACUCGCAUUAGUUGAAGGCAGCC-AGUGGCGGGGCAUCCUGUCGAGGAUGUCGAGGGUUUAGUUGCCCGUGGCAAAUGCAAUUCGCUGUAACCCCGGCAAUG .(((((((...........((((((((.((((....)))).-))).)))))(((((((....)))))))...)))))))((((((.(.((.....((.....)).....))).)))))). ( -41.20) >DroYak_CAF1 116817 120 - 1 AUAAACCCAUAAAGUUUAACUCGCAUUAGCUGAAGGCAGCCAAGUGGCGGGGCAUCCUGUCAAGGAUGUCGAGGGUUUAGUUGCCCGUGGCAAAUGCAAUUCGCUGUAACCCCGGCAAUG .(((((((...........(((((.((.((((....)))).))...)))))(((((((....)))))))...)))))))((((((.(.((.....((.....)).....))).)))))). ( -43.70) >consensus AUAAACCCAUAAAGUUUAACUCGCAUUAGUUGAAGGCAGCCAAGUGGCGGGGCAUCCUGUCAAGGAUGUCGAGGGUUUAGUUGCCCGUGGCAAAUGCAAUUCGCUGUAACCCCGGCAAUG .(((((((...........(((((.((.((((....)))).))...)))))(((((((....)))))))...)))))))((((((.(.((.....((.....)).....))).)))))). (-39.96 = -40.04 + 0.08)


| Location | 13,128,560 – 13,128,680 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.58 |
| Mean single sequence MFE | -36.02 |
| Consensus MFE | -31.04 |
| Energy contribution | -31.16 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.973031 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13128560 120 + 23771897 CUAAACCCUCGACAUCCUUAACAGGAUGUCCCGCCACUUGGCUGCCUUCAACUAAUGCGAGUUAAACUUUAUGGGUUUAUGUAAAUGAUCCCAAAAAUGUUGAUGGCACUUGGCCAGGGC .....((((.((((((((....))))))))..((((...((.((((.(((((......(((.....)))..((((.((((....)))).)))).....))))).)))))))))).)))). ( -41.30) >DroSec_CAF1 116398 120 + 1 CUAAACCCUCGACAUCCUUGACAGGAUGCCCCGCCACUUGGCUGCCUUCAACUAAUGCGAGUUAAACGUUAUGGGUUUAUGUAAAUGAUCCCAAAAAUGUUGAUGGCACUUGGCCAGGGC .....((((...((((((....))))))....((((...((.((((.(((((....(((.......)))..((((.((((....)))).)))).....))))).)))))))))).)))). ( -36.80) >DroSim_CAF1 114675 120 + 1 CUAAACCCUCGACAUCCUUGACAGGAUGCCCCGCCACUUGGCUGCCUUCAACUAAUGCGAGUUAAACUUUAUGCGUUUAUGUAAAUGAUCCCAAAAAUGUUGAUGGCACUUGGCCACGGC ............((((((....))))))....(((...((((((((.(((((..(((((((.....)))...))))((((....))))..........))))).)))....))))).))) ( -28.20) >DroEre_CAF1 113404 119 + 1 CUAAACCCUCGACAUCCUCGACAGGAUGCCCCGCCACU-GGCUGCCUUCAACUAAUGCGAGUUAAACUUUAUGGGUUUAUGUAAAUGAUCCCAAAAAUGUUGAUGGGGCUUGGCCGGGGC .............(((((....)))))(((((((((..-((((.((.(((((......(((.....)))..((((.((((....)))).)))).....))))).))))))))).)))))) ( -39.60) >DroYak_CAF1 116857 120 + 1 CUAAACCCUCGACAUCCUUGACAGGAUGCCCCGCCACUUGGCUGCCUUCAGCUAAUGCGAGUUAAACUUUAUGGGUUUAUGUAAAUGAUCCCAAAAAUGUUGAUGGUGCUUGGCCAGGAC ............((((((....))))))((..((((.(((((((....))))))).(((.(((((..(((.((((.((((....)))).)))).)))..)))))..))).))))..)).. ( -34.20) >consensus CUAAACCCUCGACAUCCUUGACAGGAUGCCCCGCCACUUGGCUGCCUUCAACUAAUGCGAGUUAAACUUUAUGGGUUUAUGUAAAUGAUCCCAAAAAUGUUGAUGGCACUUGGCCAGGGC .....((((...((((((....))))))....((((...((.((((.(((((((((....)))).......((((.((((....)))).)))).....))))).)))))))))).)))). (-31.04 = -31.16 + 0.12)


| Location | 13,128,560 – 13,128,680 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.58 |
| Mean single sequence MFE | -39.06 |
| Consensus MFE | -32.36 |
| Energy contribution | -33.00 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.935719 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13128560 120 - 23771897 GCCCUGGCCAAGUGCCAUCAACAUUUUUGGGAUCAUUUACAUAAACCCAUAAAGUUUAACUCGCAUUAGUUGAAGGCAGCCAAGUGGCGGGACAUCCUGUUAAGGAUGUCGAGGGUUUAG (((((..((...((((.(((((.(((.((((..............)))).)))((.......))....))))).))))(((....)))))((((((((....)))))))).))))).... ( -42.44) >DroSec_CAF1 116398 120 - 1 GCCCUGGCCAAGUGCCAUCAACAUUUUUGGGAUCAUUUACAUAAACCCAUAACGUUUAACUCGCAUUAGUUGAAGGCAGCCAAGUGGCGGGGCAUCCUGUCAAGGAUGUCGAGGGUUUAG (((((..((...((((.(((((.....((((..............))))...((.......)).....))))).))))(((....)))))((((((((....)))))))).))))).... ( -41.54) >DroSim_CAF1 114675 120 - 1 GCCGUGGCCAAGUGCCAUCAACAUUUUUGGGAUCAUUUACAUAAACGCAUAAAGUUUAACUCGCAUUAGUUGAAGGCAGCCAAGUGGCGGGGCAUCCUGUCAAGGAUGUCGAGGGUUUAG (((.((((....((((.(((((.....((..(....)..))(((((.......)))))..........))))).))))))))...)))..((((((((....)))))))).......... ( -36.90) >DroEre_CAF1 113404 119 - 1 GCCCCGGCCAAGCCCCAUCAACAUUUUUGGGAUCAUUUACAUAAACCCAUAAAGUUUAACUCGCAUUAGUUGAAGGCAGCC-AGUGGCGGGGCAUCCUGUCGAGGAUGUCGAGGGUUUAG ((((((((...(((((.(((((.(((.((((..............)))).)))((.......))....))))).....(((-...))))))))(((((....))))))))).)))).... ( -38.74) >DroYak_CAF1 116857 120 - 1 GUCCUGGCCAAGCACCAUCAACAUUUUUGGGAUCAUUUACAUAAACCCAUAAAGUUUAACUCGCAUUAGCUGAAGGCAGCCAAGUGGCGGGGCAUCCUGUCAAGGAUGUCGAGGGUUUAG (((((((.......)))....((....))))))........(((((((...........(((((.((.((((....)))).))...)))))(((((((....)))))))...))))))). ( -35.70) >consensus GCCCUGGCCAAGUGCCAUCAACAUUUUUGGGAUCAUUUACAUAAACCCAUAAAGUUUAACUCGCAUUAGUUGAAGGCAGCCAAGUGGCGGGGCAUCCUGUCAAGGAUGUCGAGGGUUUAG (((((.((((.(((((.(((((.....((((..............))))....((.......))....))))).))).))....))))..((((((((....)))))))).))))).... (-32.36 = -33.00 + 0.64)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:27:38 2006