
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,125,708 – 13,125,819 |
| Length | 111 |
| Max. P | 0.883467 |

| Location | 13,125,708 – 13,125,819 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.37 |
| Mean single sequence MFE | -16.18 |
| Consensus MFE | -14.04 |
| Energy contribution | -13.64 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.883467 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
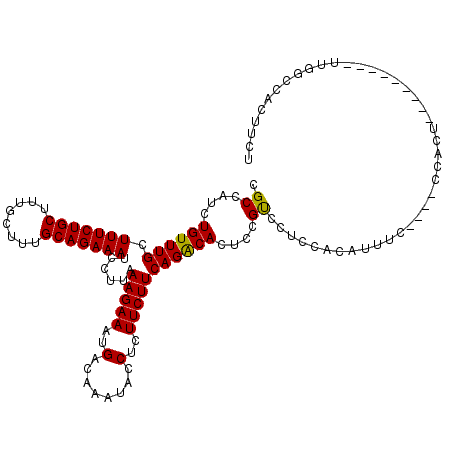
>3L_DroMel_CAF1 13125708 111 + 23771897 CGCCAUCUGUUUGCUUUCUGCUUUGCUUUGCAGAAAUCCUUAAAGAAAUGACAAAUACCUCUUCUUCAGACACUCCGUUCUCCACAUUUCCCUCCCACU---------UUUGCCACUUUU .......((((((.(((((((........)))))))......(((((..(........)..)))))))))))...........................---------............ ( -13.80) >DroSec_CAF1 113569 107 + 1 CGCCAUCUGUUUGCUUUCUGCUUUGCUUUGCAGAAAUCCUUAAAGAAAUGACAAAUACCUCUUCUUCAGACACUCCGUCCUCCACAUUUC----CCACU---------CUGGCCACUUCU .((((..((((((.(((((((........)))))))......(((((..(........)..)))))))))))....((.....)).....----.....---------.))))....... ( -18.00) >DroSim_CAF1 111805 107 + 1 CGCCAUCUGUUUGCUUUCUGCUUUGCUUUGCAGAAAUCCUUAAAGAAAUGACAAAUACCUCUUCUUCAGACACUCCGUCCUCCAUAUUUC----CCACU---------UUGGCCACUUCU .(((((((......(((((((........))))))).......)))......................(((.....)))...........----.....---------.))))....... ( -16.82) >DroEre_CAF1 110558 91 + 1 CGCCAUCUGUUUGCUUUCUGCUUAGCUUUGCAGAAAUCCUUAAAGAAAUGACAAAUACCUCUUCUUCAGGCAUUCCGCCACU-----------------------------CCCACGUCC .(((.(((((..(((........)))...)))))........(((((..(........)..)))))..)))...........-----------------------------......... ( -15.90) >DroYak_CAF1 113856 116 + 1 CGCCAGUUGUUUGCUUUCUGCUUGGCUUUGCAGAAAUCCUUAAAGAAAUGACAAAUACCUUUUCUUCAGACACUCCGCCCAUCACUUCUC----CCACUUCCCCCCGUUUUUCCACGUCC .((.(((.(((((.(((((((........)))))))......((((((.(........).))))))))))))))..))............----...........(((......)))... ( -16.40) >consensus CGCCAUCUGUUUGCUUUCUGCUUUGCUUUGCAGAAAUCCUUAAAGAAAUGACAAAUACCUCUUCUUCAGACACUCCGUCCUCCACAUUUC____CCACU_________UUGGCCACUUCU .((....((((((.(((((((........)))))))......(((((..(........)..)))))))))))....)).......................................... (-14.04 = -13.64 + -0.40)

| Location | 13,125,708 – 13,125,819 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.37 |
| Mean single sequence MFE | -24.16 |
| Consensus MFE | -17.72 |
| Energy contribution | -17.44 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.541986 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
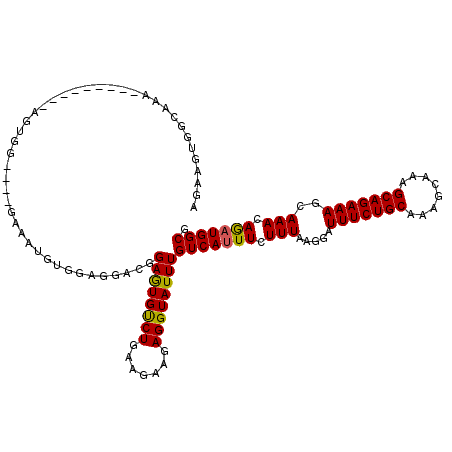
>3L_DroMel_CAF1 13125708 111 - 23771897 AAAAGUGGCAAA---------AGUGGGAGGGAAAUGUGGAGAACGGAGUGUCUGAAGAAGAGGUAUUUGUCAUUUCUUUAAGGAUUUCUGCAAAGCAAAGCAGAAAGCAAACAGAUGGCG .......((...---------.............(((.....)))....(((((..((((((((.......)))))))).....(((((((........))))))).....))))).)). ( -20.00) >DroSec_CAF1 113569 107 - 1 AGAAGUGGCCAG---------AGUGG----GAAAUGUGGAGGACGGAGUGUCUGAAGAAGAGGUAUUUGUCAUUUCUUUAAGGAUUUCUGCAAAGCAAAGCAGAAAGCAAACAGAUGGCG .......((((.---------.((..----(......((((((((((...(((.....)))....)))))).))))........(((((((........))))))).)..))...)))). ( -23.80) >DroSim_CAF1 111805 107 - 1 AGAAGUGGCCAA---------AGUGG----GAAAUAUGGAGGACGGAGUGUCUGAAGAAGAGGUAUUUGUCAUUUCUUUAAGGAUUUCUGCAAAGCAAAGCAGAAAGCAAACAGAUGGCG .......(((((---------(((((----.((((((...((((.....)))).........)))))).)))))).........(((((((........))))))).........)))). ( -23.40) >DroEre_CAF1 110558 91 - 1 GGACGUGGG-----------------------------AGUGGCGGAAUGCCUGAAGAAGAGGUAUUUGUCAUUUCUUUAAGGAUUUCUGCAAAGCUAAGCAGAAAGCAAACAGAUGGCG ...((((((-----------------------------(((((((.(((((((.......))))))))))))))))).......(((((((........)))))))...........))) ( -27.10) >DroYak_CAF1 113856 116 - 1 GGACGUGGAAAAACGGGGGGAAGUGG----GAGAAGUGAUGGGCGGAGUGUCUGAAGAAAAGGUAUUUGUCAUUUCUUUAAGGAUUUCUGCAAAGCCAAGCAGAAAGCAAACAACUGGCG .............(((..(...((.(----((((((((((.....((((..((.......))..))))))))))))))).....(((((((........)))))))))...)..)))... ( -26.50) >consensus AGAAGUGGCAAA_________AGUGG____GAAAUGUGGAGGACGGAGUGUCUGAAGAAGAGGUAUUUGUCAUUUCUUUAAGGAUUUCUGCAAAGCAAAGCAGAAAGCAAACAGAUGGCG .............................................((((((((.......))))))))(((((((.(((.....(((((((........)))))))..))).))))))). (-17.72 = -17.44 + -0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:27:26 2006