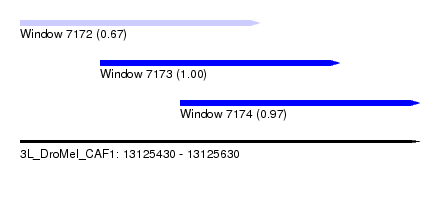
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,125,430 – 13,125,630 |
| Length | 200 |
| Max. P | 0.996806 |

| Location | 13,125,430 – 13,125,550 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -35.10 |
| Consensus MFE | -30.64 |
| Energy contribution | -32.56 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.667824 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13125430 120 + 23771897 CAUACAGUAUCUUAAAGUCGGUCUUUCCAAUCCGUUUCGUAAUAAUGUUACGUGUUUCACCUUCAUCACGUAGCCAGGCGCCUAACGGGGCCAUCCUGGAGGCCGUUGGCACCCGCCCCU ..................(((((..((((.................((((((((............))))))))..(((.((....)).)))....)))))))))..(((....)))... ( -34.40) >DroSec_CAF1 113291 120 + 1 CAUACAGUAUCUUAAAGUCGGUCUUUCCAAUCCGUUUCGUAACAAUGUUACGUGUUUCACCUUCAUCACGUAGCCAGGCGCCUAACGGGGCCAUCCUGGAGGCCGUUGGCACCCGCCCCU ..................(((((..((((.................((((((((............))))))))..(((.((....)).)))....)))))))))..(((....)))... ( -34.40) >DroSim_CAF1 111526 120 + 1 CAUACAGUAUCUUAAAGUCGGUCUUUCCAAUCCGUUUCGUAACAAUGUUACGUGUUUCACCUUCAUCACGUAGCCAGGCGCCUAACGGGGCCAUCCUGGAGGCCGUUGGCACCCGCCCCU ..................(((((..((((.................((((((((............))))))))..(((.((....)).)))....)))))))))..(((....)))... ( -34.40) >DroEre_CAF1 110282 120 + 1 CCUACAGUAUCUUAAAGUCGGCCCUUUCAAUCCGUUGCGUAAUAACAUUACGUGUUUCACCUUCAUCACGUAGCCGGGCGCCUAACGGGGCCAUCCUGGAGGCGGUAGGCACCCGCCCCU ..................((((...........((((.....))))..((((((............))))))))))(((.((....)).))).....((.(((((.......))))))). ( -35.70) >DroYak_CAF1 113579 120 + 1 CAUACAGUAUCUUAAAGUCGGCCUUUUCAAUCCGUUUCGUAAUAACGUUACGUGUUCCACCUUCAUCACGUAGCCAGGCGCCUAACGGGGCCAUCCUGGAGGCCGUUGGCACCCGCCUCU ..................((((((((....................((((((((............))))))))..(((.((....)).))).....))))))))..(((....)))... ( -36.60) >consensus CAUACAGUAUCUUAAAGUCGGUCUUUCCAAUCCGUUUCGUAAUAAUGUUACGUGUUUCACCUUCAUCACGUAGCCAGGCGCCUAACGGGGCCAUCCUGGAGGCCGUUGGCACCCGCCCCU ..................(((((..((((.................((((((((............))))))))..(((.((....)).)))....)))))))))..(((....)))... (-30.64 = -32.56 + 1.92)

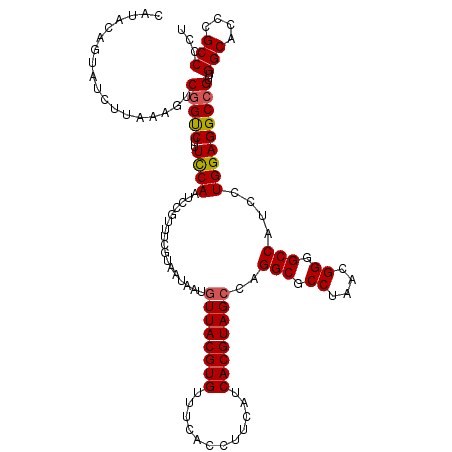
| Location | 13,125,470 – 13,125,590 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.00 |
| Mean single sequence MFE | -39.86 |
| Consensus MFE | -37.46 |
| Energy contribution | -37.34 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.75 |
| SVM RNA-class probability | 0.996806 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13125470 120 + 23771897 AAUAAUGUUACGUGUUUCACCUUCAUCACGUAGCCAGGCGCCUAACGGGGCCAUCCUGGAGGCCGUUGGCACCCGCCCCUUUCGGUUCCGUUAAAUUUGUUUAUUGCGGCACUUGAACAC ......((((((((............))))))))((((.(((((((((((((.....((.(((.(.......).)))))....)))))))))).....((.....))))).))))..... ( -38.80) >DroSec_CAF1 113331 120 + 1 AACAAUGUUACGUGUUUCACCUUCAUCACGUAGCCAGGCGCCUAACGGGGCCAUCCUGGAGGCCGUUGGCACCCGCCCCUUUCGGUUCCGUUAAAUUUGUUUAUUGCGGCACUUGAACAC ......((((((((............))))))))((((.(((((((((((((.....((.(((.(.......).)))))....)))))))))).....((.....))))).))))..... ( -38.80) >DroSim_CAF1 111566 120 + 1 AACAAUGUUACGUGUUUCACCUUCAUCACGUAGCCAGGCGCCUAACGGGGCCAUCCUGGAGGCCGUUGGCACCCGCCCCUUUCGGUUCCGUUAAAUUUGUUUAUUGCGGCACUUGAACAC ......((((((((............))))))))((((.(((((((((((((.....((.(((.(.......).)))))....)))))))))).....((.....))))).))))..... ( -38.80) >DroEre_CAF1 110322 120 + 1 AAUAACAUUACGUGUUUCACCUUCAUCACGUAGCCGGGCGCCUAACGGGGCCAUCCUGGAGGCGGUAGGCACCCGCCCCUUUCGGUUCCGUUAAAUUUGUUUAUUGCGGCACUUGAACAC .......(((((((............))))))).((((.(((((((((((((.....((.(((((.......)))))))....)))))))))).....((.....))))).))))..... ( -41.10) >DroYak_CAF1 113619 120 + 1 AAUAACGUUACGUGUUCCACCUUCAUCACGUAGCCAGGCGCCUAACGGGGCCAUCCUGGAGGCCGUUGGCACCCGCCUCUUUCGGUUCCGUUAAAUUUGUUUAUUGCGGCACUUGAACAC ......((((((((............))))))))((((.(((((((((((((.....((((((.(.......).))))))...)))))))))).....((.....))))).))))..... ( -41.80) >consensus AAUAAUGUUACGUGUUUCACCUUCAUCACGUAGCCAGGCGCCUAACGGGGCCAUCCUGGAGGCCGUUGGCACCCGCCCCUUUCGGUUCCGUUAAAUUUGUUUAUUGCGGCACUUGAACAC ......((((((((............))))))))((((.(((((((((((((.....((.(((.(.......).)))))....)))))))))).....((.....))))).))))..... (-37.46 = -37.34 + -0.12)

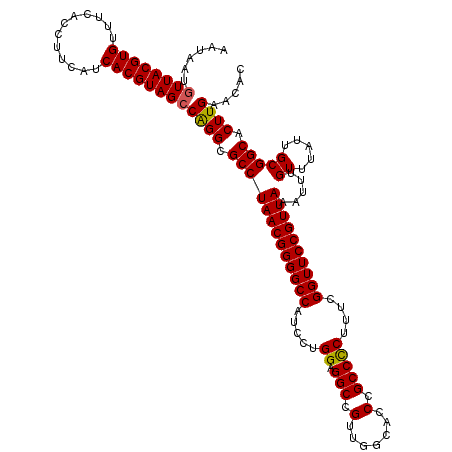
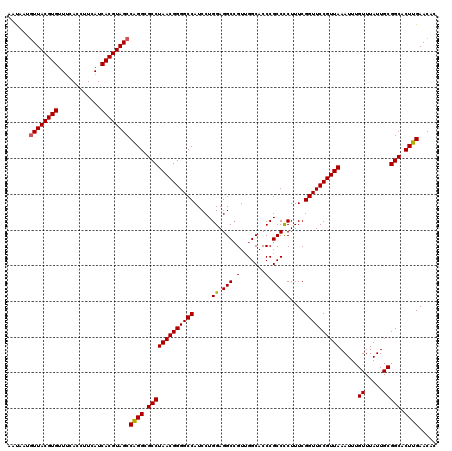
| Location | 13,125,510 – 13,125,630 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.49 |
| Mean single sequence MFE | -34.16 |
| Consensus MFE | -32.60 |
| Energy contribution | -32.44 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969595 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13125510 120 + 23771897 CCUAACGGGGCCAUCCUGGAGGCCGUUGGCACCCGCCCCUUUCGGUUCCGUUAAAUUUGUUUAUUGCGGCACUUGAACACAAGUGGCAUCAUCUUUAUAUCCCUUUCCCCCCGGAACUCC ......(((((......((..(((...)))..)))))))....(((((((........((.....))(.((((((....)))))).)........................))))))).. ( -33.60) >DroSec_CAF1 113371 119 + 1 CCUAACGGGGCCAUCCUGGAGGCCGUUGGCACCCGCCCCUUUCGGUUCCGUUAAAUUUGUUUAUUGCGGCACUUGAACACAAGUGGCAUCAUCUUCAUAUCCCUUUCC-CCCGGAACUCC ......(((((......((..(((...)))..)))))))....(((((((........((.....))(.((((((....)))))).).....................-..))))))).. ( -33.60) >DroSim_CAF1 111606 119 + 1 CCUAACGGGGCCAUCCUGGAGGCCGUUGGCACCCGCCCCUUUCGGUUCCGUUAAAUUUGUUUAUUGCGGCACUUGAACACAAGUGGCAUCAUCUUCAUAUCCCUUUCC-CCCGGAACUCC ......(((((......((..(((...)))..)))))))....(((((((........((.....))(.((((((....)))))).).....................-..))))))).. ( -33.60) >DroEre_CAF1 110362 119 + 1 CCUAACGGGGCCAUCCUGGAGGCGGUAGGCACCCGCCCCUUUCGGUUCCGUUAAAUUUGUUUAUUGCGGCACUUGAACACAAGUGGCAUCAUCUUCAUAUCCCACUCC-CCCGGAACUCC ((((((((((((.....((.(((((.......)))))))....))))))))))..............(.((((((....)))))).).....................-...))...... ( -36.70) >DroYak_CAF1 113659 119 + 1 CCUAACGGGGCCAUCCUGGAGGCCGUUGGCACCCGCCUCUUUCGGUUCCGUUAAAUUUGUUUAUUGCGGCACUUGAACACAAGUGGCAUCAUCUUCAUAUCCCAUUCC-CCCGGAACUCC ((((((((((((.....((((((.(.......).))))))...))))))))))..............(.((((((....)))))).).....................-...))...... ( -33.30) >consensus CCUAACGGGGCCAUCCUGGAGGCCGUUGGCACCCGCCCCUUUCGGUUCCGUUAAAUUUGUUUAUUGCGGCACUUGAACACAAGUGGCAUCAUCUUCAUAUCCCUUUCC_CCCGGAACUCC ......(((((......((..((.....))..)))))))....(((((((........((.....))(.((((((....)))))).)........................))))))).. (-32.60 = -32.44 + -0.16)

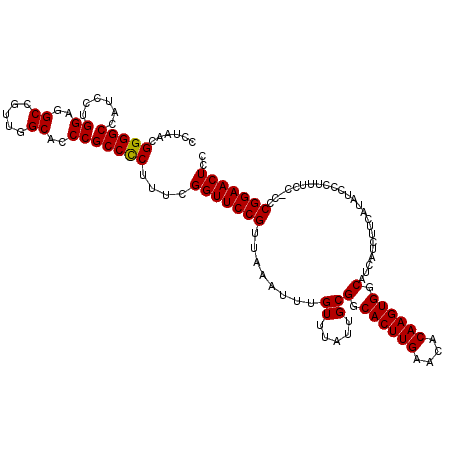
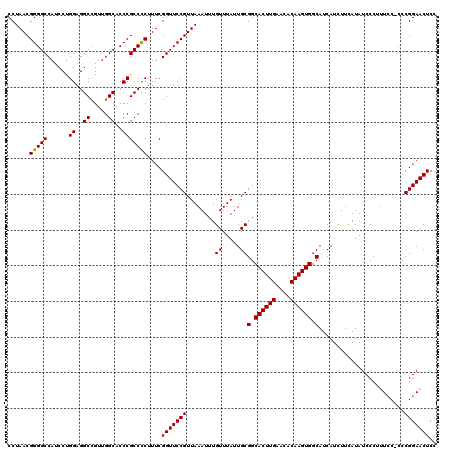
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:27:25 2006