
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,122,875 – 13,123,068 |
| Length | 193 |
| Max. P | 0.999982 |

| Location | 13,122,875 – 13,122,988 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 97.35 |
| Mean single sequence MFE | -30.00 |
| Consensus MFE | -28.60 |
| Energy contribution | -28.23 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.906407 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13122875 113 + 23771897 AUCGAAGCCUUUCAUUCCUCGGCGUGUUCGCAUUUAAAUAAUGGUCCAAGAUUAUCAAUUAAGCCGUUGGCCAAAUUAAUAGUUGGCCGACUAAAAGGACCACGCCCUAGAUA ((((((........)))...((((((....((((.....))))((((..(.....).........(((((((((........))))))))).....))))))))))...))). ( -31.00) >DroSec_CAF1 110770 113 + 1 AUCGAAGCCUUUCAUUCCUCGGCGUGUUCGCAUUUAAAUAAUGGUCAAAGAUUAUCAAUUAAGCCGUUGGCCAAAUUAAUAGUUGGCCGACUAAAAGGACCACGCCCUAGAAA ..............(((...(((((((((((..((((.(.((((((...)))))).).)))))).(((((((((........))))))))).....))).))))))...))). ( -29.50) >DroSim_CAF1 108948 113 + 1 AUCGAAGCCUUUCAUUCCUCGGCGUGUUCGCAUUUAAAUAAUGGUCAAAGAUUAUCAAUUAAGCCGUUGGCCAAAUUAAUAGUUGGCCGACUAAAAGGACCACGCCCUAGAAA ..............(((...(((((((((((..((((.(.((((((...)))))).).)))))).(((((((((........))))))))).....))).))))))...))). ( -29.50) >DroYak_CAF1 110998 113 + 1 AUCGAAGCCUUUCAUUCCUCGGCGUGUUCGCUUUUGAAUAAUGGUCAAAGAUUAUCAAUUAAGCCGUUGGCCAAAUUAAUAGUUGGUCGACUAAAAGGACCACGCCCUCGAAA ..............(((...(((((((((((((((((.((((........))))))))..)))).(((((((((........))))))))).....))).))))))...))). ( -30.00) >consensus AUCGAAGCCUUUCAUUCCUCGGCGUGUUCGCAUUUAAAUAAUGGUCAAAGAUUAUCAAUUAAGCCGUUGGCCAAAUUAAUAGUUGGCCGACUAAAAGGACCACGCCCUAGAAA .(((((........)))...(((((((((((..((((.(.((((((...)))))).).)))))).(((((((((........))))))))).....))).))))))...)).. (-28.60 = -28.23 + -0.38)

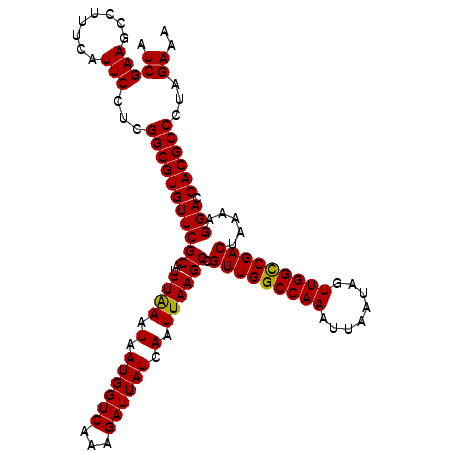
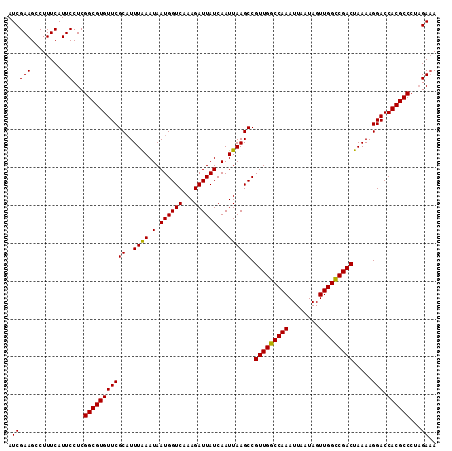
| Location | 13,122,908 – 13,123,028 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.67 |
| Mean single sequence MFE | -34.36 |
| Consensus MFE | -31.24 |
| Energy contribution | -31.64 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.90 |
| SVM RNA-class probability | 0.997658 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13122908 120 + 23771897 UUAAAUAAUGGUCCAAGAUUAUCAAUUAAGCCGUUGGCCAAAUUAAUAGUUGGCCGACUAAAAGGACCACGCCCUAGAUACCAAUGCCUCCUGGACAUGGAGUCCUGCAUGGCUGUGCUA ........((((((..(.....).........(((((((((........))))))))).....)))))).((((..(((.(((.((((....)).))))).)))..)...)))....... ( -34.90) >DroSec_CAF1 110803 120 + 1 UUAAAUAAUGGUCAAAGAUUAUCAAUUAAGCCGUUGGCCAAAUUAAUAGUUGGCCGACUAAAAGGACCACGCCCUAGAAACCAAUGCCUCUAGGACAUGGAGUCCUGCACGGCUGUGCUA .......((((((...))))))......(((((((((((((........)))))))))....(((((..((.((((((..........))))))...))..)))))....))))...... ( -35.80) >DroSim_CAF1 108981 120 + 1 UUAAAUAAUGGUCAAAGAUUAUCAAUUAAGCCGUUGGCCAAAUUAAUAGUUGGCCGACUAAAAGGACCACGCCCUAGAAACCAAUGCCUCUAGGACAUGGAGUCCUGCACGGCUGUGCUA .......((((((...))))))......(((((((((((((........)))))))))....(((((..((.((((((..........))))))...))..)))))....))))...... ( -35.80) >DroEre_CAF1 107513 120 + 1 UUGAAUAAUGGUCAAAGAUUAUCAAUUAAGCCGUUGGCCAAAUUAAUAGUUGGUCGACUAAAAGGGCCACGCCCCAGAAAGCAAUGCCUCUAGGACAUGAAGUCCUGCACGGCUGUGCUA ((((.((((........)))))))).......(((((((((........))))))))).....((((...)))).....((((..(((..((((((.....))))))...)))..)))). ( -34.80) >DroYak_CAF1 111031 120 + 1 UUGAAUAAUGGUCAAAGAUUAUCAAUUAAGCCGUUGGCCAAAUUAAUAGUUGGUCGACUAAAAGGACCACGCCCUCGAAACCAAUGCCUCUAGGACAUGGAGUCCUGCACGGCUGUGCUA ((((.((((........))))))))...(((((((((((((........)))))))))....(((((..((....))...(((.((((....)).))))).)))))....))))...... ( -30.50) >consensus UUAAAUAAUGGUCAAAGAUUAUCAAUUAAGCCGUUGGCCAAAUUAAUAGUUGGCCGACUAAAAGGACCACGCCCUAGAAACCAAUGCCUCUAGGACAUGGAGUCCUGCACGGCUGUGCUA .......((((((...))))))......(((((((((((((........)))))))))....(((((..((.((((((..........))))))...))..)))))....))))...... (-31.24 = -31.64 + 0.40)

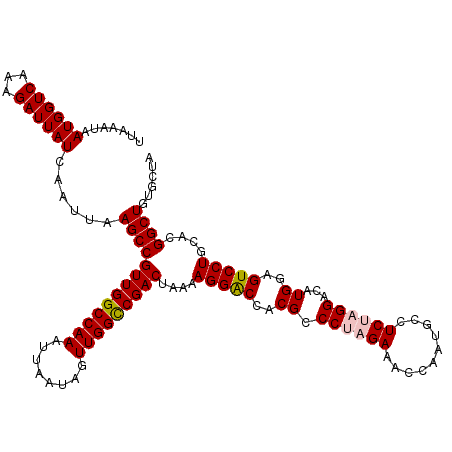

| Location | 13,122,908 – 13,123,028 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.67 |
| Mean single sequence MFE | -41.92 |
| Consensus MFE | -39.98 |
| Energy contribution | -40.74 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.70 |
| Structure conservation index | 0.95 |
| SVM decision value | 5.28 |
| SVM RNA-class probability | 0.999982 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13122908 120 - 23771897 UAGCACAGCCAUGCAGGACUCCAUGUCCAGGAGGCAUUGGUAUCUAGGGCGUGGUCCUUUUAGUCGGCCAACUAUUAAUUUGGCCAACGGCUUAAUUGAUAAUCUUGGACCAUUAUUUAA ..(((......)))((((..((((((((.(((.((....)).))).)))))))))))).......((((((.((((((((.((((...)))).))))))))...)))).))......... ( -38.10) >DroSec_CAF1 110803 120 - 1 UAGCACAGCCGUGCAGGACUCCAUGUCCUAGAGGCAUUGGUUUCUAGGGCGUGGUCCUUUUAGUCGGCCAACUAUUAAUUUGGCCAACGGCUUAAUUGAUAAUCUUUGACCAUUAUUUAA ......((((((..((((..(((((((((((((((....))))))))))))))))))).......((((((........)))))).))))))............................ ( -46.00) >DroSim_CAF1 108981 120 - 1 UAGCACAGCCGUGCAGGACUCCAUGUCCUAGAGGCAUUGGUUUCUAGGGCGUGGUCCUUUUAGUCGGCCAACUAUUAAUUUGGCCAACGGCUUAAUUGAUAAUCUUUGACCAUUAUUUAA ......((((((..((((..(((((((((((((((....))))))))))))))))))).......((((((........)))))).))))))............................ ( -46.00) >DroEre_CAF1 107513 120 - 1 UAGCACAGCCGUGCAGGACUUCAUGUCCUAGAGGCAUUGCUUUCUGGGGCGUGGCCCUUUUAGUCGACCAACUAUUAAUUUGGCCAACGGCUUAAUUGAUAAUCUUUGACCAUUAUUCAA ..((((....))))(((...(((((((((((((((...)).))))))))))))).)))....(((((.....((((((((.((((...)))).))))))))....))))).......... ( -38.30) >DroYak_CAF1 111031 120 - 1 UAGCACAGCCGUGCAGGACUCCAUGUCCUAGAGGCAUUGGUUUCGAGGGCGUGGUCCUUUUAGUCGACCAACUAUUAAUUUGGCCAACGGCUUAAUUGAUAAUCUUUGACCAUUAUUCAA ..((((....))))((((..(((((((((.(((((....))))).)))))))))))))....(((((.....((((((((.((((...)))).))))))))....))))).......... ( -41.20) >consensus UAGCACAGCCGUGCAGGACUCCAUGUCCUAGAGGCAUUGGUUUCUAGGGCGUGGUCCUUUUAGUCGGCCAACUAUUAAUUUGGCCAACGGCUUAAUUGAUAAUCUUUGACCAUUAUUUAA ..((((....))))((((..(((((((((((((((....)))))))))))))))))))....(((((.....((((((((.((((...)))).))))))))....))))).......... (-39.98 = -40.74 + 0.76)

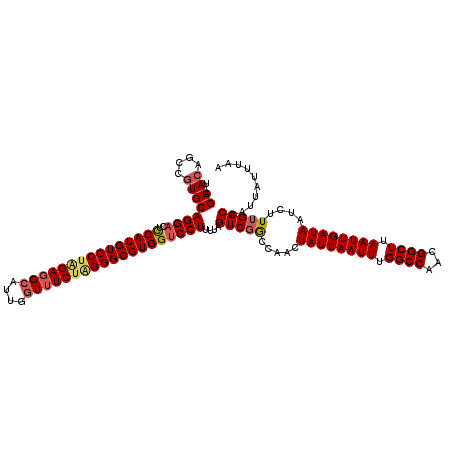
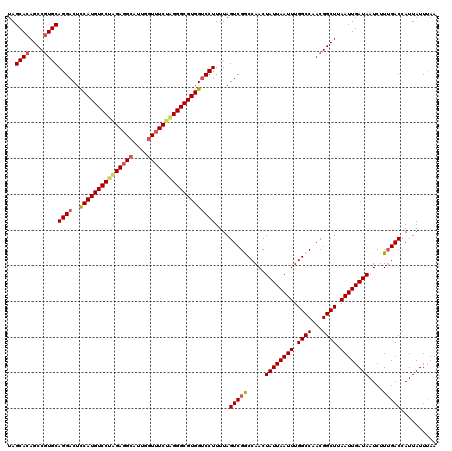
| Location | 13,122,948 – 13,123,068 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.83 |
| Mean single sequence MFE | -27.99 |
| Consensus MFE | -23.68 |
| Energy contribution | -23.44 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.817457 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13122948 120 + 23771897 AAUUAAUAGUUGGCCGACUAAAAGGACCACGCCCUAGAUACCAAUGCCUCCUGGACAUGGAGUCCUGCAUGGCUGUGCUAUCAUUAUUGUCCUCAUUAUGGCAUUAAAAUUAUUUAAUUA (((((((((((.((((......(((((.........((((.((..(((....((((.....)))).....)))..)).))))......))))).....)))).....))))).)))))). ( -26.46) >DroSec_CAF1 110843 120 + 1 AAUUAAUAGUUGGCCGACUAAAAGGACCACGCCCUAGAAACCAAUGCCUCUAGGACAUGGAGUCCUGCACGGCUGUGCUAUCAUUAUUGUCCUCAUUAUGACAUUAAAAUUAUUUAAUUA ((((((((((((((((......(((((..((.((((((..........))))))...))..)))))...)))))).))))...(((.((((........)))).)))......)))))). ( -28.80) >DroSim_CAF1 109021 120 + 1 AAUUAAUAGUUGGCCGACUAAAAGGACCACGCCCUAGAAACCAAUGCCUCUAGGACAUGGAGUCCUGCACGGCUGUGCUAUCAUUAUUGUCCUCAUUAUGGCAUUAAAAUUAUUUAAUUA ((((((((((((((((......(((((..((.((((((..........))))))...))..)))))...)))))(((((((.((..........)).)))))))...))))).)))))). ( -28.60) >DroEre_CAF1 107553 120 + 1 AAUUAAUAGUUGGUCGACUAAAAGGGCCACGCCCCAGAAAGCAAUGCCUCUAGGACAUGAAGUCCUGCACGGCUGUGCUAUCAUUACUGUUCUCAUUAUAGCAUUAAAAUUAUUUAAUUA (((((((((((....)))))...((((...))))..((.((((..(((..((((((.....))))))...)))..)))).)).(((.((((........)))).)))......)))))). ( -28.30) >DroYak_CAF1 111071 120 + 1 AAUUAAUAGUUGGUCGACUAAAAGGACCACGCCCUCGAAACCAAUGCCUCUAGGACAUGGAGUCCUGCACGGCUGUGCUAUCAUUAUUGUCCUCAUCAUGGCAUUAAAAUUAUUUAAUUA (((((((((((((((.........))))..............((((((...((((((.........((((....)))).........))))))......))))))..))))).)))))). ( -27.77) >consensus AAUUAAUAGUUGGCCGACUAAAAGGACCACGCCCUAGAAACCAAUGCCUCUAGGACAUGGAGUCCUGCACGGCUGUGCUAUCAUUAUUGUCCUCAUUAUGGCAUUAAAAUUAUUUAAUUA ((((((((((((((((......(((((..((.((((((..........))))))...))..)))))...)))))).))))...(((.((((........)))).)))......)))))). (-23.68 = -23.44 + -0.24)

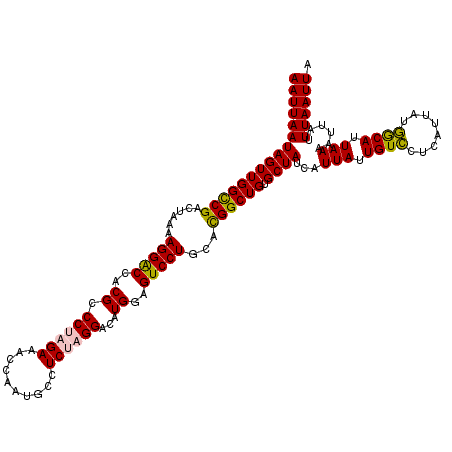
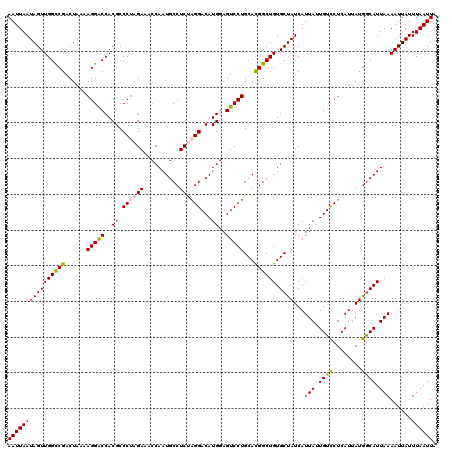
| Location | 13,122,948 – 13,123,068 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.83 |
| Mean single sequence MFE | -34.96 |
| Consensus MFE | -31.38 |
| Energy contribution | -32.38 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.90 |
| SVM decision value | 3.86 |
| SVM RNA-class probability | 0.999664 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13122948 120 - 23771897 UAAUUAAAUAAUUUUAAUGCCAUAAUGAGGACAAUAAUGAUAGCACAGCCAUGCAGGACUCCAUGUCCAGGAGGCAUUGGUAUCUAGGGCGUGGUCCUUUUAGUCGGCCAACUAUUAAUU ..((((((....))))))(((((...((((((..........(((......))).......(((((((.(((.((....)).))).)))))))))))))...)).)))............ ( -29.90) >DroSec_CAF1 110843 120 - 1 UAAUUAAAUAAUUUUAAUGUCAUAAUGAGGACAAUAAUGAUAGCACAGCCGUGCAGGACUCCAUGUCCUAGAGGCAUUGGUUUCUAGGGCGUGGUCCUUUUAGUCGGCCAACUAUUAAUU .............(((.((((........)))).)))((((((....((((.((((((..(((((((((((((((....)))))))))))))))))))....))))))...))))))... ( -40.50) >DroSim_CAF1 109021 120 - 1 UAAUUAAAUAAUUUUAAUGCCAUAAUGAGGACAAUAAUGAUAGCACAGCCGUGCAGGACUCCAUGUCCUAGAGGCAUUGGUUUCUAGGGCGUGGUCCUUUUAGUCGGCCAACUAUUAAUU ..((((((....))))))(((((...((((((..........((((....)))).......((((((((((((((....))))))))))))))))))))...)).)))............ ( -40.60) >DroEre_CAF1 107553 120 - 1 UAAUUAAAUAAUUUUAAUGCUAUAAUGAGAACAGUAAUGAUAGCACAGCCGUGCAGGACUUCAUGUCCUAGAGGCAUUGCUUUCUGGGGCGUGGCCCUUUUAGUCGACCAACUAUUAAUU ..((((((....))))))((((.....((((.(((((((...((((....))))(((((.....))))).....)))))))))))(((((...)))))..))))................ ( -30.60) >DroYak_CAF1 111071 120 - 1 UAAUUAAAUAAUUUUAAUGCCAUGAUGAGGACAAUAAUGAUAGCACAGCCGUGCAGGACUCCAUGUCCUAGAGGCAUUGGUUUCGAGGGCGUGGUCCUUUUAGUCGACCAACUAUUAAUU ..((((((....))))))....((((((((((..........((((....)))).......((((((((.(((((....))))).))))))))))))))...)))).............. ( -33.20) >consensus UAAUUAAAUAAUUUUAAUGCCAUAAUGAGGACAAUAAUGAUAGCACAGCCGUGCAGGACUCCAUGUCCUAGAGGCAUUGGUUUCUAGGGCGUGGUCCUUUUAGUCGGCCAACUAUUAAUU ..((((((....))))))...........(((..........((((....))))((((..(((((((((((((((....)))))))))))))))))))....)))............... (-31.38 = -32.38 + 1.00)


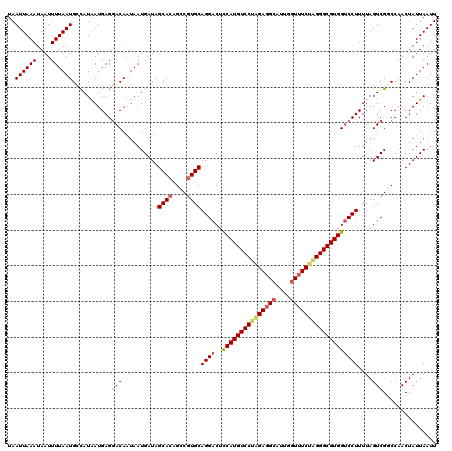
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:27:20 2006