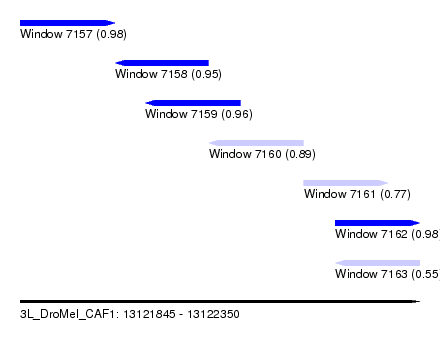
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,121,845 – 13,122,350 |
| Length | 505 |
| Max. P | 0.984911 |

| Location | 13,121,845 – 13,121,965 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.83 |
| Mean single sequence MFE | -27.78 |
| Consensus MFE | -27.60 |
| Energy contribution | -27.60 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.984911 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13121845 120 + 23771897 UGAUUUUUUGUUUACCAUUAUAGCUGAGCUGUCGUCGCCUUUCCAUUCGUCCUUUCAGCCAUUCCUCCUUGUUGGCCACGUUGAUUUACGUUGCAGGCGAACGGCAUUUCAAUUAAAUUU ....................(((.((((.(((((((((((...((..(((....(((((....((........))....)))))...))).)).))))).)))))).)))).)))..... ( -27.60) >DroSec_CAF1 109735 120 + 1 UGAUUUUUUGUUUACCAUUAUAGCUGAGCUGUCGUCGCCUUUCCAUUCGUCCUUUCAGCCAAUCCUCCUUGUUGGCCAAGUUGAUUUACGUUGCAGGCGAACGGCAUUUCAAUUAAAUUU ....................(((.((((.(((((((((((...((..(((....(((((((((.......)))))).....)))...))).)).))))).)))))).)))).)))..... ( -27.80) >DroSim_CAF1 107904 120 + 1 UGAUUUUUUGUUUACCAUUAUAGCUGAGCUGUCGUCGCCUUUCCAUUCGUCCUUUCAGCCAAUCCUCCUUGUUGGCCACGUUGAUUUACGUUGCAGGCGAACGGCAUUUCAAUUAAAUUU ....................(((.((((.(((((((((((.................((((((.......)))))).((((......))))...))))).)))))).)))).)))..... ( -28.30) >DroEre_CAF1 106435 120 + 1 UGAUUUUUUGUUUACCAUUAUAGCUGAGCUGUCGUCGCCUUUCCAUUCGUCCUUUCAGCCACUCCUCCUUGUUGGCCAAGUUGAUUUACGUUGCAGGCGAACGGCAUUUCAAUUAAAUUU ....................(((.((((.(((((((((((...((..(((....(((((....((........))....)))))...))).)).))))).)))))).)))).)))..... ( -27.60) >DroYak_CAF1 109962 120 + 1 UGAUUUUUUGUUUACCAUUAUAGCUGAGCUGUCGUCGCCUUUCCAUUCGUCCUUUCAGCCACUCCUCCUUGUUGGCCAAGUUGAUUUACGUUGCAGGCGAACGGCAUUUCAAUUAAAUUU ....................(((.((((.(((((((((((...((..(((....(((((....((........))....)))))...))).)).))))).)))))).)))).)))..... ( -27.60) >consensus UGAUUUUUUGUUUACCAUUAUAGCUGAGCUGUCGUCGCCUUUCCAUUCGUCCUUUCAGCCAAUCCUCCUUGUUGGCCAAGUUGAUUUACGUUGCAGGCGAACGGCAUUUCAAUUAAAUUU ....................(((.((((.(((((((((((...((..(((....(((((....((........))....)))))...))).)).))))).)))))).)))).)))..... (-27.60 = -27.60 + -0.00)


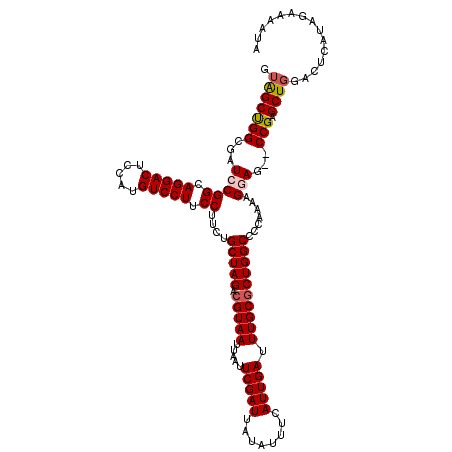
| Location | 13,121,965 – 13,122,083 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.64 |
| Mean single sequence MFE | -36.86 |
| Consensus MFE | -30.14 |
| Energy contribution | -30.62 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.950173 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13121965 118 - 23771897 GUAGCUGGCGAUCCGGCAGGACUCCAUGUCCUUCCUUCUGCUAGACGUAAUUAAUUCGAUUAUAUUUCAUUGAUUUGCGCUGGCCCCAAAAGGAG--CCGAGCUGGACUCAUAGAAAAUA (.(((..((....(((((((((.....)))))(((((..(((((.(((((.....(((((........))))).)))))))))).....))))))--))).))..).))).......... ( -36.60) >DroSec_CAF1 109855 120 - 1 GUAGCUGGCGAUCCGGCAGGACUCCGUGUCCUUCCUUCUGCUAGACGUAAUUAAUUCGAUUAUAUUUCAUUGAUUUGCUCUGGCCCCAAAAGUAGCACCGAGCUGGACUCAUAGAAUAUA ....(((..(((((((((((((.....))))).......((((((.((((.....(((((........))))).)))))))))).................)))))).)).)))...... ( -30.10) >DroSim_CAF1 108024 120 - 1 AUAGCUGGCGAUCCGGCAGGACUCCAUGUCCUUCCUUCUGCUAGACGUAAUUAAUUCGAUUAUAUUUCAUUGAUUUGCGCUGGCCCCAAAAGGAGCGCCGAGCUGGACUCAUAGAAAAUA .(((((((((.(((((.(((((.....))))).))....(((((.(((((.....(((((........))))).)))))))))).......))).)))).)))))............... ( -41.10) >DroEre_CAF1 106555 118 - 1 GAGGCCGGCGAUUCGGCAGGACUCCAUGUCCUUCCUUCUGCUAGACGUAAUUAAUUCGAUUAUAUUUCAUUGAUUUGCGCUGGCCCCAAAAGCAG--CCGAGCUGGACUCAUAGAAAAUA (((.(((((...((((((((((.....))))).......(((((.(((((.....(((((........))))).))))))))))..........)--))))))))).))).......... ( -39.90) >DroYak_CAF1 110082 118 - 1 GUAGCUGGCGAUCCGGCAGGACUCCAUGUCCUUCCUUCUGCUAGACGUAAUUAAUUCGAUUAUAUUUCAUUGAUUUGCGCUGGCCCCAAAAGGAG--CCGAGCUGGACUCAUAGAAAAUA (.(((..((....(((((((((.....)))))(((((..(((((.(((((.....(((((........))))).)))))))))).....))))))--))).))..).))).......... ( -36.60) >consensus GUAGCUGGCGAUCCGGCAGGACUCCAUGUCCUUCCUUCUGCUAGACGUAAUUAAUUCGAUUAUAUUUCAUUGAUUUGCGCUGGCCCCAAAAGGAG__CCGAGCUGGACUCAUAGAAAAUA .(((((((...(((((.(((((.....))))).))....(((((.(((((.....(((((........))))).)))))))))).......)))...))).))))............... (-30.14 = -30.62 + 0.48)


| Location | 13,122,003 – 13,122,123 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.67 |
| Mean single sequence MFE | -32.44 |
| Consensus MFE | -31.26 |
| Energy contribution | -31.66 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958246 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13122003 120 - 23771897 CAAUCGAAUCGCAAAUUUAUGUGGCACCUAUACAAGGAGCGUAGCUGGCGAUCCGGCAGGACUCCAUGUCCUUCCUUCUGCUAGACGUAAUUAAUUCGAUUAUAUUUCAUUGAUUUGCGC .........((((((((.(((..((.(((.....))).))...((((((((...((.(((((.....))))).)).)).))))).)((((((.....))))))....))).)))))))). ( -33.00) >DroSec_CAF1 109895 120 - 1 CAAUCGAAUCGCAAAUUUAUGUGGCACCUAUACAAGGAGCGUAGCUGGCGAUCCGGCAGGACUCCGUGUCCUUCCUUCUGCUAGACGUAAUUAAUUCGAUUAUAUUUCAUUGAUUUGCUC ..........(((((((.(((..((.(((.....))).))...((((((((...((.(((((.....))))).)).)).))))).)((((((.....))))))....))).))))))).. ( -31.60) >DroSim_CAF1 108064 120 - 1 CAAUCGAAUCGCAAAUUUAUGUGGCACCUAUACAAGGAGCAUAGCUGGCGAUCCGGCAGGACUCCAUGUCCUUCCUUCUGCUAGACGUAAUUAAUUCGAUUAUAUUUCAUUGAUUUGCGC .........((((((((.(((..((.(((.....))).))...((((((((...((.(((((.....))))).)).)).))))).)((((((.....))))))....))).)))))))). ( -33.00) >DroEre_CAF1 106593 120 - 1 CAAUCGAAUCGCAAAUUUAUGUGGCACCUAUACAAGGAGCGAGGCCGGCGAUUCGGCAGGACUCCAUGUCCUUCCUUCUGCUAGACGUAAUUAAUUCGAUUAUAUUUCAUUGAUUUGCGC .........((((((((.((((((((.......(((((.....(((((....)))))(((((.....)))))))))).)))))...((((((.....))))))....))).)))))))). ( -31.60) >DroYak_CAF1 110120 120 - 1 CAAUCGAAUCGCAAAUUUAUGUGGCACCUAUACAAGGAGCGUAGCUGGCGAUCCGGCAGGACUCCAUGUCCUUCCUUCUGCUAGACGUAAUUAAUUCGAUUAUAUUUCAUUGAUUUGCGC .........((((((((.(((..((.(((.....))).))...((((((((...((.(((((.....))))).)).)).))))).)((((((.....))))))....))).)))))))). ( -33.00) >consensus CAAUCGAAUCGCAAAUUUAUGUGGCACCUAUACAAGGAGCGUAGCUGGCGAUCCGGCAGGACUCCAUGUCCUUCCUUCUGCUAGACGUAAUUAAUUCGAUUAUAUUUCAUUGAUUUGCGC .........((((((((.(((..((.(((.....))).))...((((((((...((.(((((.....))))).)).)).))))).)((((((.....))))))....))).)))))))). (-31.26 = -31.66 + 0.40)

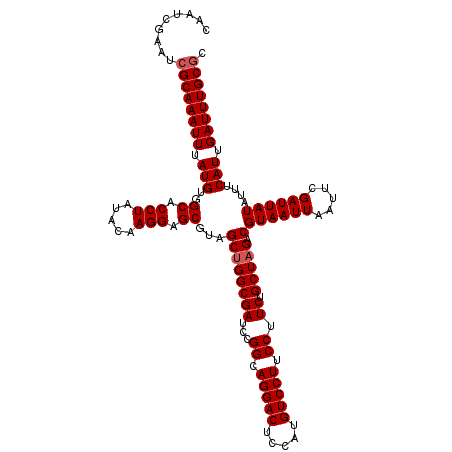
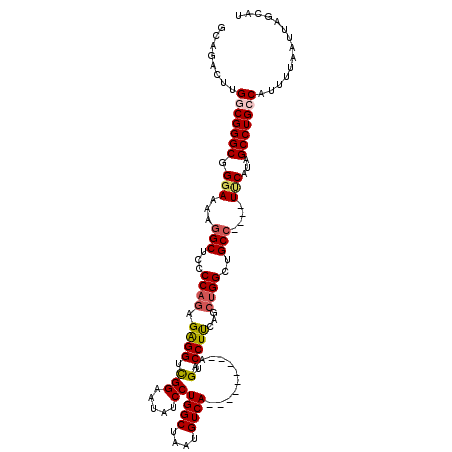
| Location | 13,122,083 – 13,122,203 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.42 |
| Mean single sequence MFE | -27.54 |
| Consensus MFE | -26.70 |
| Energy contribution | -26.54 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.887503 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13122083 120 - 23771897 CGAAUACCGCUGCUGGCCAAUACUGAAUUUGGCCAUUAACAAUAAUCCGAAAUCCAUAGAAAUCUGUGGCCAAUGAAACGCAAUCGAAUCGCAAAUUUAUGUGGCACCUAUACAAGGAGC ........((...(((((((........)))))))..........(((......(((((....)))))(((((((((..((.........))...))))).))))..........))))) ( -27.00) >DroSec_CAF1 109975 120 - 1 CGAAUACCGCUUCUGGCCAAUACUGAAUUUGGCCAUUAACAAUAAUCCGAAAUCCAUAGAAAUCUGUGGCCAAUGAAACGCAAUCGAAUCGCAAAUUUAUGUGGCACCUAUACAAGGAGC ........((((((((((((........))))))....................(((((....)))))(((((((((..((.........))...))))).)))).........)))))) ( -28.50) >DroSim_CAF1 108144 120 - 1 CGAAUACCGCUGCUGGCCAAUACUGAAUUUGGCCAUUAACAAUAAUCCGAAAUCCAUAGAAAUCUGUGGCCAAUGAAACGCAAUCGAAUCGCAAAUUUAUGUGGCACCUAUACAAGGAGC ........((...(((((((........)))))))..........(((......(((((....)))))(((((((((..((.........))...))))).))))..........))))) ( -27.00) >DroEre_CAF1 106673 120 - 1 CGCAUACCGCUGCUGGCCAAUACCGAAUUUGGCUAUUAACAAUAAUCCGAAAUCCAUAGAAAUCUGUGGCCAAUGAAACGCAAUCGAAUCGCAAAUUUAUGUGGCACCUAUACAAGGAGC ((((((....((((((((((........)))))))............(((...((((((....))))))....((.....)).)))....)))....))))))((.(((.....))).)) ( -27.70) >DroYak_CAF1 110200 120 - 1 CGAAUACCGCUGCUGGCCAAUACGGAAUUUGGCCAUUAACAAUAAUCCGAAAUCCAUAGAAAUCUGUGGCCAAUGAAACGCAAUCGAAUCGCAAAUUUAUGUGGCACCUAUACAAGGAGC ........((..((((((.....(((.(((((..((((....))))))))).)))((((....))))))))).......((.........))........)..)).(((.....)))... ( -27.50) >consensus CGAAUACCGCUGCUGGCCAAUACUGAAUUUGGCCAUUAACAAUAAUCCGAAAUCCAUAGAAAUCUGUGGCCAAUGAAACGCAAUCGAAUCGCAAAUUUAUGUGGCACCUAUACAAGGAGC ........((...(((((((........)))))))..........(((......(((((....)))))(((((((((..((.........))...))))).))))..........))))) (-26.70 = -26.54 + -0.16)


| Location | 13,122,203 – 13,122,310 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.30 |
| Mean single sequence MFE | -37.89 |
| Consensus MFE | -29.86 |
| Energy contribution | -30.46 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.766479 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13122203 107 + 23771897 ACAGACUUGGCGGGCGGGAAAAAGCUCCCCAGAGAGGAUGGAAUAUCCUGGCUAAUGUCA---------AAGUCCUUCAGAUGGCUGCC----UUCAUAGCCUGCCAUUUUAAUUAGCAU ...((..((((((((..(((...((...(((..((((((((.....))((((....))))---------..))))))....)))..)).----)))...))))))))..))......... ( -33.40) >DroSec_CAF1 110095 107 + 1 GCAGACUUGGCGGGCGGGAAAAGGCUCCCCAGAGAGGUCGGAAUAUCCUGGCUAAUGUCA---------AAGUCCUUCAGCUGGCUGCC----UUCAUAGCCUGCCAUUUUAAUUAGCAU ...((..((((((((..((..((((...((((.((((.(((.....))((((....))))---------..).))))...))))..)))----)))...))))))))..))......... ( -39.20) >DroSim_CAF1 108264 116 + 1 GCAGACUUGGCGGGCGGGAAAAGGCUCCCCAGAGAGGUCGGAAUAUCCUGGCUAAUGUCAGGAAAAAAAAAGUCCUUCAGCUGGCUGCC----UUCAUAGCCUGCCAUUUUAAUUAGCAU ...((..((((((((..((..((((...((((.((((.(......(((((((....)))))))........).))))...))))..)))----)))...))))))))..))......... ( -44.54) >DroEre_CAF1 106793 107 + 1 GCAGACUUGCCGGGCGGGAAAAGGCUCCCCAGAGAGGUCGGAAUAUCCUGGCUGAUGUCA---------AAGUCCUCUAGCCGGCUGCC----UCCGUAGCCUGCCUUUUUAAUUAGCAU .......(((..(....((((((((.......(((((.(((.....))((((....))))---------..).)))))....((((((.----...)))))).))))))))..)..))). ( -35.70) >DroYak_CAF1 110320 111 + 1 GCAGACUUGGCGGGCGGGAAAAGGCUCCCCAGAGGGGUCGGAAUAUCCUGGCUAAUGUCA---------AAGUCCUUUAGCUGGCUGCCUGCCUACAUAGCCUGGCCUUUUAAUUAGCAU .(((.((.(((((((((.....((((((.....))))))........(..(((((.....---------.......)))))..)))))))))).....)).)))................ ( -36.60) >consensus GCAGACUUGGCGGGCGGGAAAAGGCUCCCCAGAGAGGUCGGAAUAUCCUGGCUAAUGUCA_________AAGUCCUUCAGCUGGCUGCC____UUCAUAGCCUGCCAUUUUAAUUAGCAU ........(((((((.(((...(((...((((.((((.(((.....))((((....))))...........).))))...))))..)))....)))...))))))).............. (-29.86 = -30.46 + 0.60)


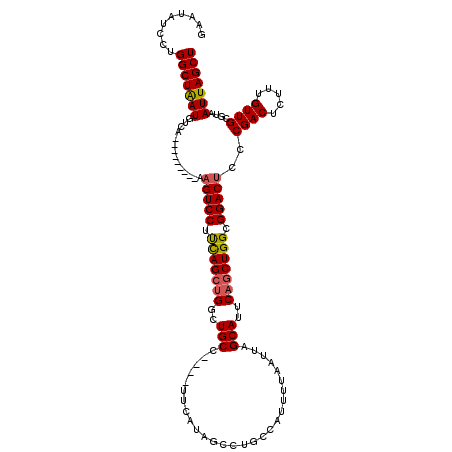
| Location | 13,122,243 – 13,122,350 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.55 |
| Mean single sequence MFE | -31.58 |
| Consensus MFE | -26.75 |
| Energy contribution | -26.79 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.977612 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13122243 107 + 23771897 GAAUAUCCUGGCUAAUGUCA---------AAGUCCUUCAGAUGGCUGCC----UUCAUAGCCUGCCAUUUUAAUUAGCAUUCAGCUGGCGGACUCCCGACUCUUUGUUGCGUAAUUAGCU .........(((((((....---------.(((((...(((((((.((.----......))..)))))))...(((((.....))))).)))))..((((.....))))....))))))) ( -27.40) >DroSec_CAF1 110135 107 + 1 GAAUAUCCUGGCUAAUGUCA---------AAGUCCUUCAGCUGGCUGCC----UUCAUAGCCUGCCAUUUUAAUUAGCAUUCAGCUGGCGGACCACCGACUCUUUGUUGCGUAAUUAGCU .........(((((((....---------..((((.(((((((((((..----....)))))(((...........)))...)))))).)))).((((((.....)))).)).))))))) ( -30.70) >DroSim_CAF1 108304 116 + 1 GAAUAUCCUGGCUAAUGUCAGGAAAAAAAAAGUCCUUCAGCUGGCUGCC----UUCAUAGCCUGCCAUUUUAAUUAGCAUUCAGCUGGCGGACUACCGACUCUUUGUUGCGUAAUUAGCU .....(((((((....)))))))........((((.(((((((((((..----....)))))(((...........)))...)))))).))))(((((((.....)))).)))....... ( -36.70) >DroEre_CAF1 106833 107 + 1 GAAUAUCCUGGCUGAUGUCA---------AAGUCCUCUAGCCGGCUGCC----UCCGUAGCCUGCCUUUUUAAUUAGCAUUCAGCUGGCGGACUCCCGACUCUUUGUUGCGUAAUUAGCU .........(((((((....---------.(((((.(((((.((((((.----...))))))(((...........)))....))))).)))))..((((.....))))....))))))) ( -32.70) >DroYak_CAF1 110360 111 + 1 GAAUAUCCUGGCUAAUGUCA---------AAGUCCUUUAGCUGGCUGCCUGCCUACAUAGCCUGGCCUUUUAAUUAGCAUUCAGCUGGCGGACUCCCGACUCUUUGUUGCGUAAUUAGCU .........(((((((....---------.(((((.(((((((..(((..(((..........)))..........)))..))))))).)))))..((((.....))))....))))))) ( -30.40) >consensus GAAUAUCCUGGCUAAUGUCA_________AAGUCCUUCAGCUGGCUGCC____UUCAUAGCCUGCCAUUUUAAUUAGCAUUCAGCUGGCGGACUCCCGACUCUUUGUUGCGUAAUUAGCU .........(((((((..............(((((.(((((((..(((............................)))..))))))).)))))..((((.....))))....))))))) (-26.75 = -26.79 + 0.04)


| Location | 13,122,243 – 13,122,350 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.55 |
| Mean single sequence MFE | -30.54 |
| Consensus MFE | -25.42 |
| Energy contribution | -27.10 |
| Covariance contribution | 1.68 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.550163 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13122243 107 - 23771897 AGCUAAUUACGCAACAAAGAGUCGGGAGUCCGCCAGCUGAAUGCUAAUUAAAAUGGCAGGCUAUGAA----GGCAGCCAUCUGAAGGACUU---------UGACAUUAGCCAGGAUAUUC .((((((..(........).(((((.(((((((((....(((....)))....)))).((((.....----...)))).......))))))---------)))))))))).......... ( -29.00) >DroSec_CAF1 110135 107 - 1 AGCUAAUUACGCAACAAAGAGUCGGUGGUCCGCCAGCUGAAUGCUAAUUAAAAUGGCAGGCUAUGAA----GGCAGCCAGCUGAAGGACUU---------UGACAUUAGCCAGGAUAUUC .((((((..(........).(((((.(((((..((((((..((((..((...((((....)))).))----))))..))))))..))))))---------)))))))))).......... ( -32.70) >DroSim_CAF1 108304 116 - 1 AGCUAAUUACGCAACAAAGAGUCGGUAGUCCGCCAGCUGAAUGCUAAUUAAAAUGGCAGGCUAUGAA----GGCAGCCAGCUGAAGGACUUUUUUUUUCCUGACAUUAGCCAGGAUAUUC .((((((..(........).(((((.(((((..((((((..((((..((...((((....)))).))----))))..))))))..))))).........))))))))))).......... ( -32.10) >DroEre_CAF1 106833 107 - 1 AGCUAAUUACGCAACAAAGAGUCGGGAGUCCGCCAGCUGAAUGCUAAUUAAAAAGGCAGGCUACGGA----GGCAGCCGGCUAGAGGACUU---------UGACAUCAGCCAGGAUAUUC .((.......))......(((((((.(((((.(.((((...((((.........))))((((.(...----.).)))))))).).))))))---------)))).))............. ( -27.80) >DroYak_CAF1 110360 111 - 1 AGCUAAUUACGCAACAAAGAGUCGGGAGUCCGCCAGCUGAAUGCUAAUUAAAAGGCCAGGCUAUGUAGGCAGGCAGCCAGCUAAAGGACUU---------UGACAUUAGCCAGGAUAUUC .((((((..(........).(((((.(((((...(((((..((((..(((...(((...)))...)))...))))..)))))...))))))---------)))))))))).......... ( -31.10) >consensus AGCUAAUUACGCAACAAAGAGUCGGGAGUCCGCCAGCUGAAUGCUAAUUAAAAUGGCAGGCUAUGAA____GGCAGCCAGCUGAAGGACUU_________UGACAUUAGCCAGGAUAUUC .((((((..(........).((((.((((((..((((((..((((.......((((....)))).......))))..))))))..)))))).........)))))))))).......... (-25.42 = -27.10 + 1.68)

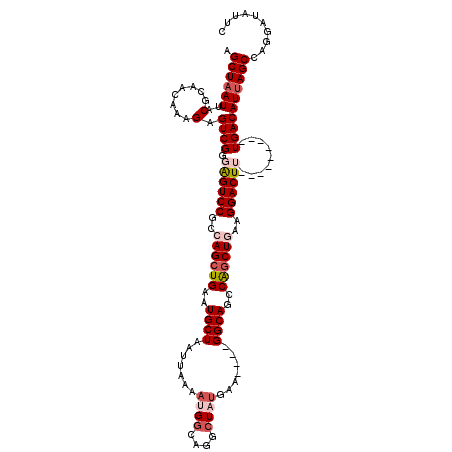
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:27:14 2006