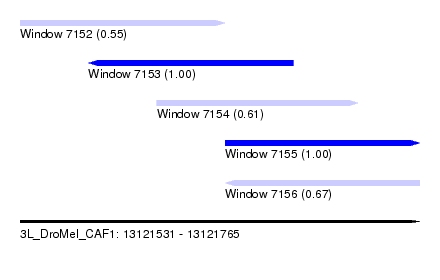
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,121,531 – 13,121,765 |
| Length | 234 |
| Max. P | 0.999551 |

| Location | 13,121,531 – 13,121,651 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.95 |
| Mean single sequence MFE | -28.76 |
| Consensus MFE | -26.20 |
| Energy contribution | -26.24 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.554732 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13121531 120 + 23771897 UGCAGGCUAUAUGGUGAAAUAUAAAAACAUUAUUAAUCAAUGUCCAGGAAAACCGGGGCAGGACACAAAAGCUGGAAGUUGGUAGUAAGUAAGCUUACUGUUGCCAUGUCUUUGACGGGA ..((((..((((((..(..................((((((.(((((.....((......)).(......)))))).))))))((((((....)))))).)..)))))).))))...... ( -26.70) >DroSec_CAF1 109436 118 + 1 UGCAGGCUAUAUGGCGAAAUAUAAAAACAUUAUUAAUCAAUGUCCAGGAAAAGCGGGGCAGGACACAAAAGCUGGAAGUUGGUAGUAAGUAAGCUUACUGUUGCCAUGUCUUUGACGG-- ..((((..(((((((((..................((((((.(((((.....((...)).(....).....))))).))))))((((((....)))))).))))))))).))))....-- ( -28.50) >DroSim_CAF1 107604 118 + 1 UGCAGGCUAUAUGGCGAAAUAUAAAAACAUUAUUAAUCAAUGUCCAGGAAAAGCGGGGCAGGACACAAAAGCUGGAAGUUGGUAGUAAGUAAGCUUACUGUUGCCAUGUCUUUGACGG-- ..((((..(((((((((..................((((((.(((((.....((...)).(....).....))))).))))))((((((....)))))).))))))))).))))....-- ( -28.50) >DroEre_CAF1 106131 118 + 1 UGCAGGCUAUAUGGCCAAAUAUAAAAACAUUAUUAAUCAAUGUCCAGGAAAAGCAGGGCAGGGCACGAAAGCUGGAAGUUGGUAGUAAGUAAGCUUACUGUUGCCAUGUCUUUGGCGG-- ....((((....)))).........(((.......((((((.(((((.....((........)).(....)))))).))))))((((((....)))))))))((((......))))..-- ( -32.60) >DroYak_CAF1 109655 113 + 1 UGCAGGCUAUAUGGCGAAAUAUAAAAACAUUAUUAAUCAAUGUCCAGGAAAAGCAG-----GGCACAAAAGCUGGAAGUUGGUAGUAAGUAAGCUUACUGUUGCCAUGUCUUUGACGG-- ..((((..(((((((((..................((((((.(((((.........-----..........))))).))))))((((((....)))))).))))))))).))))....-- ( -27.51) >consensus UGCAGGCUAUAUGGCGAAAUAUAAAAACAUUAUUAAUCAAUGUCCAGGAAAAGCGGGGCAGGACACAAAAGCUGGAAGUUGGUAGUAAGUAAGCUUACUGUUGCCAUGUCUUUGACGG__ ..((((..(((((((((..................((((((.(((((........................))))).))))))((((((....)))))).))))))))).))))...... (-26.20 = -26.24 + 0.04)


| Location | 13,121,571 – 13,121,691 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.25 |
| Mean single sequence MFE | -42.74 |
| Consensus MFE | -36.87 |
| Energy contribution | -36.75 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.86 |
| SVM decision value | 3.71 |
| SVM RNA-class probability | 0.999551 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13121571 120 - 23771897 GUCCGGGAAUCCCCCAAGGGGCGCGAAAUUGGGACAGCCGUCCCGUCAAAGACAUGGCAACAGUAAGCUUACUUACUACCAACUUCCAGCUUUUGUGUCCUGCCCCGGUUUUCCUGGACA (((((((((..((....(((((((((((((((((..(((((...(((...))))))))...((((((....))))))......)))))).))))))))))).....))..))))))))). ( -51.60) >DroSec_CAF1 109476 117 - 1 GUC-GGGAAUCCCCCAAGGGGUGCGAAAUUGGGACAGCCG--CCGUCAAAGACAUGGCAACAGUAAGCUUACUUACUACCAACUUCCAGCUUUUGUGUCCUGCCCCGCUUUUCCUGGACA (((-(((((........((((..(((((((((((..((((--..(((...))).))))...((((((....))))))......)))))).)))))..)))).........))))).))). ( -37.23) >DroSim_CAF1 107644 117 - 1 GUC-GGGAAUCCCCCAAGGGGCGCGAAAUUGGGACACCCG--CCGUCAAAGACAUGGCAACAGUAAGCUUACUUACUACCAACUUCCAGCUUUUGUGUCCUGCCCCGCUUUUCCUGGACA (((-(((((........(((((((((((((((((.....(--((((.......)))))...((((((....))))))......)))))).))))))))))).........))))).))). ( -39.43) >DroEre_CAF1 106171 117 - 1 CUC-GGGAAUCCCCCAAGGGGCGCGAAAUUGGGACAGCCG--CCGCCAAAGACAUGGCAACAGUAAGCUUACUUACUACCAACUUCCAGCUUUCGUGCCCUGCCCUGCUUUUCCUGGACA .((-(((((........(((((((((((((((((......--..((((......))))...((((((....))))))......)))))).))))))))))).........)))))))... ( -43.63) >DroYak_CAF1 109695 112 - 1 GUC-GGGAAUCCCCCAAGGGGCGCGAAAUUGGGACAGCCG--CCGUCAAAGACAUGGCAACAGUAAGCUUACUUACUACCAACUUCCAGCUUUUGUGCC-----CUGCUUUUCCUGGACA (((-(((.....)))..(((((((((((((((((..((((--..(((...))).))))...((((((....))))))......)))))).)))))))))-----))..........))). ( -41.80) >consensus GUC_GGGAAUCCCCCAAGGGGCGCGAAAUUGGGACAGCCG__CCGUCAAAGACAUGGCAACAGUAAGCUUACUUACUACCAACUUCCAGCUUUUGUGUCCUGCCCCGCUUUUCCUGGACA (((.(((((........(((((((((((((((((..........((((......))))...((((((....))))))......)))))).))))))))))).........))))).))). (-36.87 = -36.75 + -0.12)

| Location | 13,121,611 – 13,121,729 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.66 |
| Mean single sequence MFE | -43.28 |
| Consensus MFE | -28.08 |
| Energy contribution | -29.44 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.611177 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13121611 118 + 23771897 GGUAGUAAGUAAGCUUACUGUUGCCAUGUCUUUGACGGGACGGCUGUCCCAAUUUCGCGCCCCUUGGGGGAUUCCCGGACGAUUGCUAAAGCUGGAAAAAGGG-GAUCCCCC-AAAACAA (((((((((....))))))...((...(((...)))(((((....)))))......)))))..(((((((((((((........((....))........)))-))))))))-))..... ( -46.79) >DroSec_CAF1 109516 103 + 1 GGUAGUAAGUAAGCUUACUGUUGCCAUGUCUUUGACGG--CGGCUGUCCCAAUUUCGCACCCCUUGGGGGAUUCCC-GACGAUUGCUGG------AAAAAGGG-GAUCCCC-------AA (((((((((....))))))((((((..(((...)))))--))))..............))).....((((((((((-............------.....)))-)))))))-------.. ( -35.83) >DroSim_CAF1 107684 108 + 1 GGUAGUAAGUAAGCUUACUGUUGCCAUGUCUUUGACGG--CGGGUGUCCCAAUUUCGCGCCCCUUGGGGGAUUCCC-GACGAUUGCUAAAGCGGCAAAAAGGG-GAUCCCC-------A- ..(((((((....)))))))..(((..(((...)))))--)((((((.........))))))....((((((((((-.....(((((.....)))))...)))-)))))))-------.- ( -43.60) >DroEre_CAF1 106211 113 + 1 GGUAGUAAGUAAGCUUACUGUUGCCAUGUCUUUGGCGG--CGGCUGUCCCAAUUUCGCGCCCCUUGGGGGAUUCCC-GAGGAUAGCUAACGCUGGAGGAAGGG-GGUGCCCCAAAAG--- ((.((((((....))))))(((((((......))))))--)(((...(((..((((...(((((((((.....)))-))))..(((....))))).))))..)-)).))))).....--- ( -48.50) >DroYak_CAF1 109730 116 + 1 GGUAGUAAGUAAGCUUACUGUUGCCAUGUCUUUGACGG--CGGCUGUCCCAAUUUCGCGCCCCUUGGGGGAUUCCC-GACGAUAGCUAAAGCUGGAAAAAGGGGGGUUCCCCCAAAGCA- ((.((((((....))))))((((((..(((...)))))--))))....))........((...(((((((((((((-.....((((....))))......)))))..)))))))).)).- ( -41.70) >consensus GGUAGUAAGUAAGCUUACUGUUGCCAUGUCUUUGACGG__CGGCUGUCCCAAUUUCGCGCCCCUUGGGGGAUUCCC_GACGAUUGCUAAAGCUGGAAAAAGGG_GAUCCCCC_AAA__A_ (((((((((....))))))...(((.((((...))))....)))..............)))....(((((((((((......((((....))))......))).))))))))........ (-28.08 = -29.44 + 1.36)

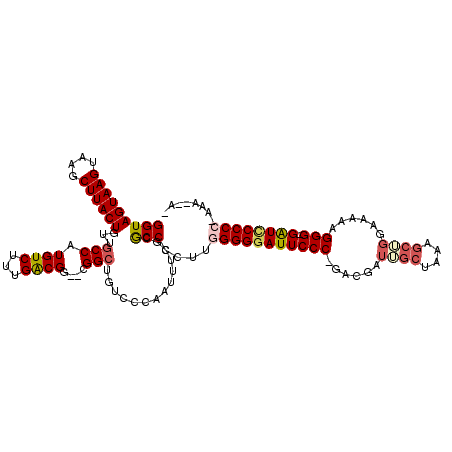
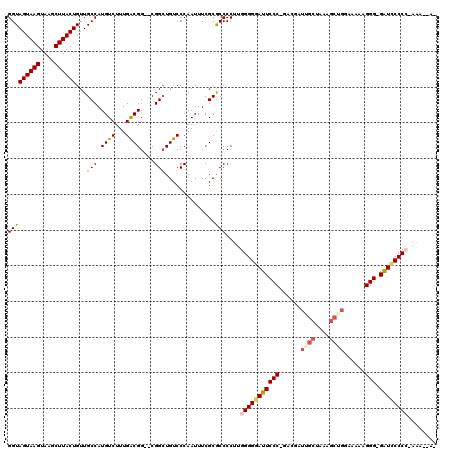
| Location | 13,121,651 – 13,121,765 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.17 |
| Mean single sequence MFE | -45.60 |
| Consensus MFE | -32.64 |
| Energy contribution | -34.16 |
| Covariance contribution | 1.52 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.04 |
| Structure conservation index | 0.72 |
| SVM decision value | 2.66 |
| SVM RNA-class probability | 0.996175 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13121651 114 + 23771897 CGGCUGUCCCAAUUUCGCGCCCCUUGGGGGAUUCCCGGACGAUUGCUAAAGCUGGAAAAAGGG-GAUCCCCC-AAAACAAAAAAAAAAAAUGGGGAUGCUG---AGCCACCAGCCGGC-A ((((((.......(((((((((((((((((((((((........((....))........)))-))))))))-))................)))).))).)---))....))))))..-. ( -46.28) >DroSec_CAF1 109554 101 + 1 CGGCUGUCCCAAUUUCGCACCCCUUGGGGGAUUCCC-GACGAUUGCUGG------AAAAAGGG-GAUCCCC-------AACAAAAAAAAAUGGGGAUGCUG---AGCCACCAGCCGGC-A ((((((.......((((((((((...((((((((((-............------.....)))-)))))))-------.............)))).))).)---))....))))))..-. ( -38.92) >DroSim_CAF1 107722 102 + 1 CGGGUGUCCCAAUUUCGCGCCCCUUGGGGGAUUCCC-GACGAUUGCUAAAGCGGCAAAAAGGG-GAUCCCC-------A-----AAAAAAUGGGGAUGCUG---AGCCACCAGCCGGC-A .(((...)))......(((((((((.((((((((((-.....(((((.....)))))...)))-)))))))-------.-----....)).)))).)))..---.(((.......)))-. ( -41.80) >DroEre_CAF1 106249 107 + 1 CGGCUGUCCCAAUUUCGCGCCCCUUGGGGGAUUCCC-GAGGAUAGCUAACGCUGGAGGAAGGG-GGUGCCCCAAAAG---------GAAUUGGGGAUGCUGUUG-GCCACCAGCCGGC-A ((((((..(((((...(((((((((((((.((((((-.....((((....))))......)))-))).))))))...---------.....)))).))).))))-)....))))))..-. ( -51.40) >DroYak_CAF1 109768 114 + 1 CGGCUGUCCCAAUUUCGCGCCCCUUGGGGGAUUCCC-GACGAUAGCUAAAGCUGGAAAAAGGGGGGUUCCCCCAAAGCA-----AAAAAUUGGGGAUGCUGCUGGGCCACCAGCCGGCGA ..(((((((((((((...((...(((((((((((((-.....((((....))))......)))))..)))))))).)).-----..))))))))).....(((((....))))))))).. ( -49.60) >consensus CGGCUGUCCCAAUUUCGCGCCCCUUGGGGGAUUCCC_GACGAUUGCUAAAGCUGGAAAAAGGG_GAUCCCCC_AAA__A_____AAAAAAUGGGGAUGCUG___AGCCACCAGCCGGC_A ((((((..........(((((((..(((((((((((......((((....))))......))).))))))))...................)))).)))...........)))))).... (-32.64 = -34.16 + 1.52)

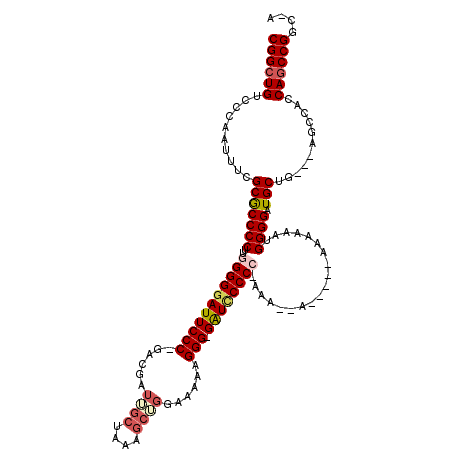
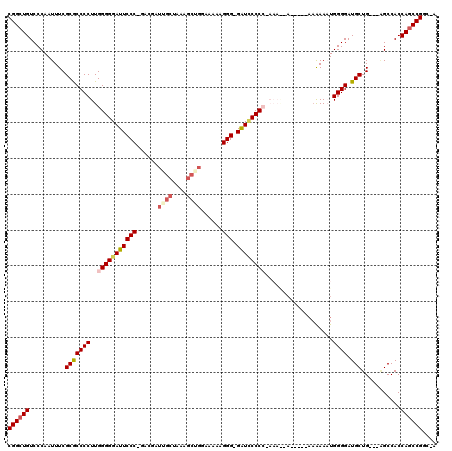
| Location | 13,121,651 – 13,121,765 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.17 |
| Mean single sequence MFE | -41.29 |
| Consensus MFE | -27.08 |
| Energy contribution | -27.52 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.666009 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13121651 114 - 23771897 U-GCCGGCUGGUGGCU---CAGCAUCCCCAUUUUUUUUUUUUGUUUU-GGGGGAUC-CCCUUUUUCCAGCUUUAGCAAUCGUCCGGGAAUCCCCCAAGGGGCGCGAAAUUGGGACAGCCG .-..((((((....((---((((..((((................((-(((((((.-(((........((....))........))).))))))))))))).)).....)))).)))))) ( -40.58) >DroSec_CAF1 109554 101 - 1 U-GCCGGCUGGUGGCU---CAGCAUCCCCAUUUUUUUUUGUU-------GGGGAUC-CCCUUUUU------CCAGCAAUCGUC-GGGAAUCCCCCAAGGGGUGCGAAAUUGGGACAGCCG .-..((((((....((---(((((((((.........(((((-------((..(..-.....)..------))))))).....-(((.....)))..))))))).....)))).)))))) ( -40.10) >DroSim_CAF1 107722 102 - 1 U-GCCGGCUGGUGGCU---CAGCAUCCCCAUUUUUU-----U-------GGGGAUC-CCCUUUUUGCCGCUUUAGCAAUCGUC-GGGAAUCCCCCAAGGGGCGCGAAAUUGGGACACCCG .-.......((((.((---(((.(((((((......-----)-------))))))(-(((((.((((.......)))).....-(((.....))))))))).......))))).)))).. ( -36.10) >DroEre_CAF1 106249 107 - 1 U-GCCGGCUGGUGGC-CAACAGCAUCCCCAAUUC---------CUUUUGGGGCACC-CCCUUCCUCCAGCGUUAGCUAUCCUC-GGGAAUCCCCCAAGGGGCGCGAAAUUGGGACAGCCG .-..((((((...((-.....))...((((((((---------(((((((((....-(((.......(((....)))......-)))....)))))))))).....))))))).)))))) ( -41.62) >DroYak_CAF1 109768 114 - 1 UCGCCGGCUGGUGGCCCAGCAGCAUCCCCAAUUUUU-----UGCUUUGGGGGAACCCCCCUUUUUCCAGCUUUAGCUAUCGUC-GGGAAUCCCCCAAGGGGCGCGAAAUUGGGACAGCCG ....((((((((.((...)).))...(((((((((.-----((((((((((((..(((.........(((....)))......-)))..))))))))..)))).))))))))).)))))) ( -48.06) >consensus U_GCCGGCUGGUGGCU___CAGCAUCCCCAUUUUUU_____U__UUU_GGGGGAUC_CCCUUUUUCCAGCUUUAGCAAUCGUC_GGGAAUCCCCCAAGGGGCGCGAAAUUGGGACAGCCG ....((((((...((......)).((((.(((((...............(((.....)))..............((..((......))..((((...)))).))))))).)))))))))) (-27.08 = -27.52 + 0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:27:07 2006