
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,120,814 – 13,120,991 |
| Length | 177 |
| Max. P | 0.993311 |

| Location | 13,120,814 – 13,120,923 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.47 |
| Mean single sequence MFE | -25.68 |
| Consensus MFE | -15.32 |
| Energy contribution | -15.32 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.60 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.511464 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
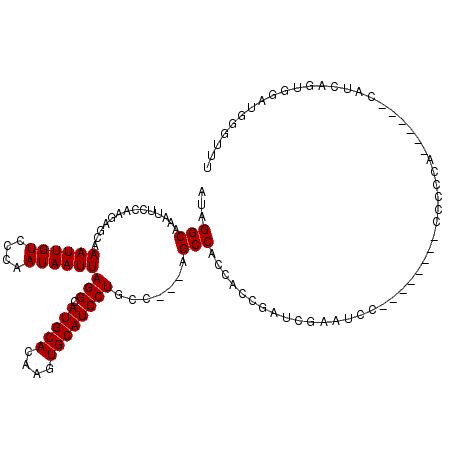
>3L_DroMel_CAF1 13120814 109 - 23771897 AUAGGCAAAUUCCAAGAGCAAAUUGUCCCAAUAAUUAGGCAUGCACAAGUGCAUCCUGCC---AGCCACCACCGAUCGAAUAC--------CCCCCAUUCCACCAACAGUGGAUGGGUUU ...(((..............((((((....)))))).((((((((....))))...))))---.)))................--------..(((((.((((.....)))))))))... ( -28.60) >DroSec_CAF1 108760 103 - 1 AUAGGCAAAUUCCAAGGGCAAAUUGUCCCAAUAAUUAGGCAUGCACAAGUGCAUCCUGCC---AGCCACCACCGAUCGAAUCC--------CCUCC------UCAUCAGUGGAUGGGUUU ...(((.........((((.....))))........(((.(((((....)))))))))))---..(((((((.(((.((....--------.....------))))).)))).))).... ( -30.00) >DroSim_CAF1 106904 103 - 1 AUAGGCAAAUUCCAAGAGCAAAUUGUCCCAAUAAUUAGGCAUGCACAAGUGCAUCCUGCC---AGCCACCACCGAUCGAAUCC--------CCUCC------UCAUCAGUGGAUGGGUUU ...(((..............((((((....))))))(((.(((((....)))))))))))---..(((((((.(((.((....--------.....------))))).)))).))).... ( -25.90) >DroEre_CAF1 105467 97 - 1 AUAGGCAAAUUCCAAGAGCAAAUUGUCCCAAUAAUUAGGCAUGCACAAGUGCAUCCUGCC--CAGCCACCACCCA---------------CCACCCA------CCGCACUGGAUGGAUUG ...(((..............((((((....)))))).((((((((....))))...))))--..)))........---------------(((.(((------......))).))).... ( -21.50) >DroYak_CAF1 108907 116 - 1 AUAGGCAAAUUCCAAGAGCAAAUUGUCCCAAUAAUUAGGCAUGCACAAGUGCAUCCUGCCAGCCGCCACCACCGAUCGAAUGCCUCCCCCCCCCCCACAA---CCCCACA-AUCGGAUGG ...(((..............((((((....)))))).((((((((....))))...))))....))).((((((((........................---.......-))))).))) ( -22.38) >consensus AUAGGCAAAUUCCAAGAGCAAAUUGUCCCAAUAAUUAGGCAUGCACAAGUGCAUCCUGCC___AGCCACCACCGAUCGAAUCC________CCCCCA______CAUCAGUGGAUGGGUUU ...(((..............((((((....))))))(((.(((((....)))))))).......)))..................................................... (-15.32 = -15.32 + 0.00)

| Location | 13,120,846 – 13,120,956 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.18 |
| Mean single sequence MFE | -38.62 |
| Consensus MFE | -33.90 |
| Energy contribution | -33.90 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.39 |
| SVM RNA-class probability | 0.993311 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13120846 110 + 23771897 UUCGAUCGGUGGUGGCU---GGCAGGAUGCACUUGUGCAUGCCUAAUUAUUGGGACAAUUUGCUCUUGGAAUUUGCCUAUUAAUUAGCCAUUGCACA-------CGGCAGACGAAGGCGA ((((.((.((.((((((---(((.(((((((....))))).))((((((.((((.(((.((.(....).)).))))))).))))))))))..)).))-------).)).))))))..... ( -38.60) >DroSec_CAF1 108786 110 + 1 UUCGAUCGGUGGUGGCU---GGCAGGAUGCACUUGUGCAUGCCUAAUUAUUGGGACAAUUUGCCCUUGGAAUUUGCCUAUUAAUUAGCCAUUGCACA-------CGGCAGACGAAGGCGA ((((.((.((.((((((---(((.(((((((....))))).))((((((.((((.(((....((...))...))))))).))))))))))..)).))-------).)).))))))..... ( -37.50) >DroSim_CAF1 106930 110 + 1 UUCGAUCGGUGGUGGCU---GGCAGGAUGCACUUGUGCAUGCCUAAUUAUUGGGACAAUUUGCUCUUGGAAUUUGCCUAUUAAUUAGCCAUUGCACA-------CGGCAGACGAAGGCGA ((((.((.((.((((((---(((.(((((((....))))).))((((((.((((.(((.((.(....).)).))))))).))))))))))..)).))-------).)).))))))..... ( -38.60) >DroEre_CAF1 105491 106 + 1 -----UGGGUGGUGGCUG--GGCAGGAUGCACUUGUGCAUGCCUAAUUAUUGGGACAAUUUGCUCUUGGAAUUUGCCUAUUAAUUAGCCAUUGCACA-------CGGCAGACGAAGGCGG -----...(..(((((((--((((...((((....)))))))))(((((.((((.(((.((.(....).)).))))))).)))))))))))..)...-------((.(.......).)). ( -35.70) >DroYak_CAF1 108943 120 + 1 UUCGAUCGGUGGUGGCGGCUGGCAGGAUGCACUUGUGCAUGCCUAAUUAUUGGGACAAUUUGCUCUUGGAAUUUGCCUAUUAAUUAGCCAUUGCACACGGCACACGGCAGACGAAGGCGA ((((.((.((.(((...((((((.....))...((((((((.(((((((.((((.(((.((.(....).)).))))))).))))))).)).)))))))))).))).)).))))))..... ( -42.70) >consensus UUCGAUCGGUGGUGGCU___GGCAGGAUGCACUUGUGCAUGCCUAAUUAUUGGGACAAUUUGCUCUUGGAAUUUGCCUAUUAAUUAGCCAUUGCACA_______CGGCAGACGAAGGCGA ........(..(((((....((((...((((....))))))))((((((.((((.(((.((.(....).)).))))))).)))))))))))..)..........((.(.......).)). (-33.90 = -33.90 + -0.00)



| Location | 13,120,883 – 13,120,991 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.41 |
| Mean single sequence MFE | -34.86 |
| Consensus MFE | -30.62 |
| Energy contribution | -30.22 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.88 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.529549 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13120883 108 + 23771897 GCCUAAUUAUUGGGACAAUUUGCUCUUGGAAUUUGCCUAUUAAUUAGCCAUUGCACA-------CGGCAGACGAAGGCGAUUCGGUCGAGUUUGCAUUUGCGG-----GCAGCCUUUGGA (((((((((.((((.(((.((.(....).)).))))))).))))))((....))...-------.)))...((((((((((...)))..((((((....))))-----)).))))))).. ( -34.10) >DroSec_CAF1 108823 107 + 1 GCCUAAUUAUUGGGACAAUUUGCCCUUGGAAUUUGCCUAUUAAUUAGCCAUUGCACA-------CGGCAGACGAAGGCGAUUCGGUCGAGUUUGCAUUUGCG------GCAGCCUUUGGA (((((((((.((((.(((....((...))...))))))).))))))((....))...-------.)))...((((((((((((....)))))(((....)))------...))))))).. ( -30.50) >DroSim_CAF1 106967 107 + 1 GCCUAAUUAUUGGGACAAUUUGCUCUUGGAAUUUGCCUAUUAAUUAGCCAUUGCACA-------CGGCAGACGAAGGCGAUUCGGUCGAGUUUGCAUUUGAG------GCAGCCUUUGGA (((((((((.((((.(((.((.(....).)).))))))).))))))((....))...-------.)))...((((((((((((....)))))(((.......------)))))))))).. ( -31.60) >DroEre_CAF1 105524 109 + 1 GCCUAAUUAUUGGGACAAUUUGCUCUUGGAAUUUGCCUAUUAAUUAGCCAUUGCACA-------CGGCAGACGAAGGCGGUUCGGUCGAGCCUGAGCCUGUGCCU----UUGCCUUUGGA ..(((((((.((((.(((.((.(....).)).))))))).)))))))(((((((...-------..)))).((((((((((((((......)))))))...))))----)))....))). ( -36.10) >DroYak_CAF1 108983 120 + 1 GCCUAAUUAUUGGGACAAUUUGCUCUUGGAAUUUGCCUAUUAAUUAGCCAUUGCACACGGCACACGGCAGACGAAGGCGAUUCGGUCGAGUUUGCCUUUGUGGCGGCGGCAGCCUUCGGA (((((((((.((((.(((.((.(....).)).))))))).))))))(((..(((.....)))...)))..(((((((((((((....))).)))))))))))))(((....)))...... ( -42.00) >consensus GCCUAAUUAUUGGGACAAUUUGCUCUUGGAAUUUGCCUAUUAAUUAGCCAUUGCACA_______CGGCAGACGAAGGCGAUUCGGUCGAGUUUGCAUUUGCG______GCAGCCUUUGGA (((((((((.((((.(((.((.(....).)).))))))).))))))((....))...........)))...((((((((((((....)))))(((.............)))))))))).. (-30.62 = -30.22 + -0.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:27:01 2006