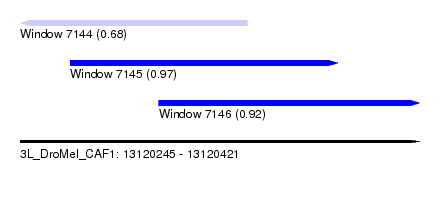
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,120,245 – 13,120,421 |
| Length | 176 |
| Max. P | 0.974533 |

| Location | 13,120,245 – 13,120,345 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.87 |
| Mean single sequence MFE | -27.90 |
| Consensus MFE | -20.98 |
| Energy contribution | -21.14 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.683973 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13120245 100 - 23771897 AAUGGGGCCAAAAGGGUUCA-ACUGUGAGCCGCAUUUGCUGGCAAAGGGAAUACGAA-UCCCAAUUAUGACGUUCAAAGUGCU------------------GUGCAUACUAAGCCAUAAA .((((.((((....((((((-....))))))((....))))))...((((.......-))))................((((.------------------..))))......))))... ( -25.20) >DroSec_CAF1 108171 100 - 1 AAUGGGGCCAAAAGGGUUCA-ACUGUGAGCCGCAUUUGCUGGCAAAGGGAAUACGAA-UCCCAAUUAUGAUGUUCAAAGUGUU------------------GAGCAUACUAAGCCAUAAA .((((.((((....((((((-....))))))((....))))))...((((.......-)))).......((((((((....))------------------))))))......))))... ( -30.50) >DroSim_CAF1 106322 100 - 1 AAUGGAGCCAAAAGGGUUCA-ACUGUGAGCCGCAUUUGCUGGCAAAGGGAAUACGAA-UCCCAAUUAUGAUGCUCAAAGUGCU------------------GUGCAUACUAAGCCAUAAA .((((.((((....((((((-....))))))((....))))))...((((.......-)))).......((((.((......)------------------).))))......))))... ( -28.20) >DroEre_CAF1 104918 96 - 1 AAUGGGGCCAAAAGGGUUCU-ACUGUGAGCCGCAUUUGCUGGCAAAGGGAAUACGAA-CCCCAAUUAUGAUGUUCAAAGUACUU-------------------GCAUACUAAGCUAU--- ..(((((((....))(((((-..(((.(((.......))).)))...))))).....-)))))..((((..((.......))..-------------------.)))).........--- ( -21.10) >DroYak_CAF1 108288 120 - 1 AAUGGGGCCAAAAGGGUUCAAACUGUGAGCCGCAUUUGCUGGCAAAGGGAAUUCGUAUUCCCAAUUAUGAUGUUCAAAGUGCUCCGCAUACUGAACAUACUGAGCAUACUAAGCUAAAAA ......((((....((((((.....))))))((....))))))...((((((....)))))).......(((((((..((((...))))..)))))))....(((.......)))..... ( -34.50) >consensus AAUGGGGCCAAAAGGGUUCA_ACUGUGAGCCGCAUUUGCUGGCAAAGGGAAUACGAA_UCCCAAUUAUGAUGUUCAAAGUGCU__________________GAGCAUACUAAGCCAUAAA .((((.((((....(((((.......)))))((....))))))...((((........))))...................................................))))... (-20.98 = -21.14 + 0.16)

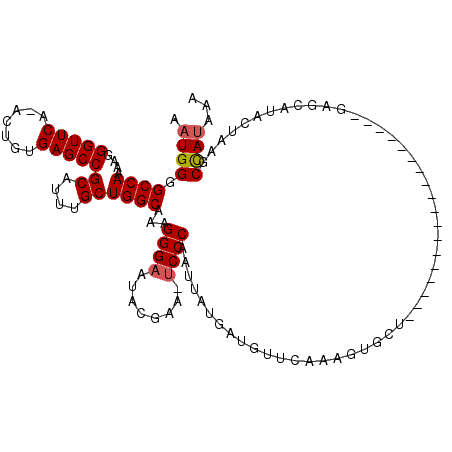
| Location | 13,120,267 – 13,120,385 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.97 |
| Mean single sequence MFE | -28.53 |
| Consensus MFE | -25.04 |
| Energy contribution | -25.08 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974533 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13120267 118 + 23771897 ACUUUGAACGUCAUAAUUGGGA-UUCGUAUUCCCUUUGCCAGCAAAUGCGGCUCACAGU-UGAACCCUUUUGGCCCCAUUAGGCUGCCACUCACAGCAGUUUAAUAUCAAUUACAAGCAA ...((((..((((.....(((.-((((..........(((.((....))))).......-)))))))...))))...((((((((((........)))))))))).)))).......... ( -27.43) >DroSec_CAF1 108193 118 + 1 ACUUUGAACAUCAUAAUUGGGA-UUCGUAUUCCCUUUGCCAGCAAAUGCGGCUCACAGU-UGAACCCUUUUGGCCCCAUUAGGCUGCCACUCACAGCAGUUUAAUAUCAAUUACAAGCAA ....(((...........((((-.......))))...(((.((....)))))))).(((-(((..((....))....((((((((((........)))))))))).))))))........ ( -26.70) >DroSim_CAF1 106344 118 + 1 ACUUUGAGCAUCAUAAUUGGGA-UUCGUAUUCCCUUUGCCAGCAAAUGCGGCUCACAGU-UGAACCCUUUUGGCUCCAUUAGGCUGCCACUCACAGCAGUUUAAUAUCAAUUACAAGCAA ....(((((.........((((-.......)))).......((....)).))))).(((-(((..((....))....((((((((((........)))))))))).))))))........ ( -28.90) >DroEre_CAF1 104936 118 + 1 ACUUUGAACAUCAUAAUUGGGG-UUCGUAUUCCCUUUGCCAGCAAAUGCGGCUCACAGU-AGAACCCUUUUGGCCCCAUUAGGCUGCCACUCACAGCAGUUUAAUAUCAAUUACAAGCAA .............(((((((((-(((.((((......(((.((....)))))....)))-)))))))..........((((((((((........))))))))))..))))))....... ( -30.10) >DroYak_CAF1 108328 120 + 1 ACUUUGAACAUCAUAAUUGGGAAUACGAAUUCCCUUUGCCAGCAAAUGCGGCUCACAGUUUGAACCCUUUUGGCCCCAUUAGGCUGCCACUCACAGCAGUUUAAUAUCAAUUACAAGCAA ....(((...........((((((....))))))...(((.((....))))))))..(((((...((....))....((((((((((........))))))))))........))))).. ( -29.50) >consensus ACUUUGAACAUCAUAAUUGGGA_UUCGUAUUCCCUUUGCCAGCAAAUGCGGCUCACAGU_UGAACCCUUUUGGCCCCAUUAGGCUGCCACUCACAGCAGUUUAAUAUCAAUUACAAGCAA ...((((...........((((........))))...((((((....))((.(((.....))).))....))))...((((((((((........)))))))))).)))).......... (-25.04 = -25.08 + 0.04)


| Location | 13,120,306 – 13,120,421 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.02 |
| Mean single sequence MFE | -24.20 |
| Consensus MFE | -17.60 |
| Energy contribution | -17.12 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922601 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13120306 115 + 23771897 AGCAAAUGCGGCUCACAGU-UGAACCCUUUUGGCCCCAUUAGGCUGCCACUCACAGCAGUUUAAUAUCAAUUACAAGCAACUAGGCCCACGCCCACACACACCU---ACAAC-CAAACAU .((....))((((...(((-((.......((((....((((((((((........)))))))))).)))).......))))).)))).................---.....-....... ( -22.54) >DroSec_CAF1 108232 116 + 1 AGCAAAUGCGGCUCACAGU-UGAACCCUUUUGGCCCCAUUAGGCUGCCACUCACAGCAGUUUAAUAUCAAUUACAAGCAACUAGGCCCACGCCCACACACACCU---ACACCCCACGCAU .....((((((((...(((-(((..((....))....((((((((((........)))))))))).))))))...))).....(((....)))...........---........))))) ( -27.00) >DroSim_CAF1 106383 116 + 1 AGCAAAUGCGGCUCACAGU-UGAACCCUUUUGGCUCCAUUAGGCUGCCACUCACAGCAGUUUAAUAUCAAUUACAAGCAACUAGGCCCACGCCCACACACACCU---ACACCCCACGCAU .....((((((((...(((-(((..((....))....((((((((((........)))))))))).))))))...))).....(((....)))...........---........))))) ( -27.00) >DroEre_CAF1 104975 103 + 1 AGCAAAUGCGGCUCACAGU-AGAACCCUUUUGGCCCCAUUAGGCUGCCACUCACAGCAGUUUAAUAUCAAUUACAAGCAACUAGG------CCCACACA---------CAC-CUACACAU .((....))........((-((.........((((..((((((((((........))))))))))..........((...)).))------))......---------...-)))).... ( -19.57) >DroYak_CAF1 108368 119 + 1 AGCAAAUGCGGCUCACAGUUUGAACCCUUUUGGCCCCAUUAGGCUGCCACUCACAGCAGUUUAAUAUCAAUUACAAGCAACUAUGGCCAAGCCAACACAUACCCCAUCCAC-CUGCACAU ......(((((......(((((...((....))....((((((((((........))))))))))........))))).....((((...)))).................-)))))... ( -24.90) >consensus AGCAAAUGCGGCUCACAGU_UGAACCCUUUUGGCCCCAUUAGGCUGCCACUCACAGCAGUUUAAUAUCAAUUACAAGCAACUAGGCCCACGCCCACACACACCU___ACACCCCACACAU ......(((((((.(.((........)).).))))..((((((((((........))))))))))...........)))....((.......)).......................... (-17.60 = -17.12 + -0.48)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:26:57 2006