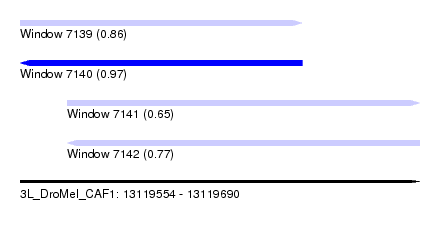
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,119,554 – 13,119,690 |
| Length | 136 |
| Max. P | 0.972425 |

| Location | 13,119,554 – 13,119,650 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.43 |
| Mean single sequence MFE | -25.27 |
| Consensus MFE | -23.12 |
| Energy contribution | -23.16 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.863003 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13119554 96 + 23771897 UCAG------------------------UCUGUCAUUUGCUAAACAACAGCAAAGGACACUGCACAUAAGUAGAAACAAACUAUACUAAUUGCCAGUUUGUCUCGCCCACGAAGGUGAAG ....------------------------..((((.((((((.......)))))).))))((((......))))..(((((((............))))))).(((((......))))).. ( -22.30) >DroSec_CAF1 107447 109 + 1 UCAGUCU---AGUCUA--------UCAGUCUGUCAUUUGCUAAACAACAGCAAAGGACACUGCACAUAAGUAGAAACAAACUAAACUAAUUGCCAGUUUGUCUCGCCCACGAAGGUGGAG ..(((.(---(((...--------......((((.((((((.......)))))).))))((((......))))......)))).)))....((.((.....)).))((((....)))).. ( -25.40) >DroSim_CAF1 105598 112 + 1 UCAGUCUAUCAGUCUA--------UCAGUCUGUCAUUUGCUAAACAACAGCAAAGGACACUGCACAUAAGUAGAAACAAACUAAACUAAUUGCCAGUUUGUCUCGCCCACGAAGGUGGAG ....(((((.......--------.((((..(((.((((((.......)))))).))))))).......))))).(((((((............))))))).....((((....)))).. ( -25.36) >DroEre_CAF1 104276 104 + 1 UCAGUCUAUCA----------------GUCUGUCAUUUGCUAAACAACGGCAAAGGACACUGCACAUAAGUAGAAACAAACUAAACUAAUUGCCAGUUUGUCUCGCCCACGAAGGUGGAG .........((----------------((..(((.((((((.......)))))).)))))))..........((.(((((((............))))))).))..((((....)))).. ( -25.00) >DroYak_CAF1 107569 120 + 1 UCAGUCUAUCAGUCCAUCAGUCCGUCAGUCUGUCAUUUGCUAAACAACAGCAAAGGACACUGCACAUAAGUAGAAACAAACUAAACUAAUUGCCAGUUUGUCUCGCCCACGAAGGUGGAG ............((((((.....((((((..(((.((((((.......)))))).))))))).)).......((.(((((((............))))))).)).........)))))). ( -28.30) >consensus UCAGUCUAUCAGUC_A________UCAGUCUGUCAUUUGCUAAACAACAGCAAAGGACACUGCACAUAAGUAGAAACAAACUAAACUAAUUGCCAGUUUGUCUCGCCCACGAAGGUGGAG ..............................((((.((((((.......)))))).))))((((......))))..(((((((............))))))).....((((....)))).. (-23.12 = -23.16 + 0.04)


| Location | 13,119,554 – 13,119,650 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.43 |
| Mean single sequence MFE | -31.34 |
| Consensus MFE | -28.60 |
| Energy contribution | -28.64 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.972425 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13119554 96 - 23771897 CUUCACCUUCGUGGGCGAGACAAACUGGCAAUUAGUAUAGUUUGUUUCUACUUAUGUGCAGUGUCCUUUGCUGUUGUUUAGCAAAUGACAGA------------------------CUGA .........((((((.(((((((((((((.....)).)))))))))))..))))))..(((((((.(((((((.....))))))).)))..)------------------------))). ( -29.20) >DroSec_CAF1 107447 109 - 1 CUCCACCUUCGUGGGCGAGACAAACUGGCAAUUAGUUUAGUUUGUUUCUACUUAUGUGCAGUGUCCUUUGCUGUUGUUUAGCAAAUGACAGACUGA--------UAGACU---AGACUGA ..((((....))))((.((.....)).))..((((((((((((((...(((....)))(((((((.(((((((.....))))))).)))..)))))--------))))))---))))))) ( -35.10) >DroSim_CAF1 105598 112 - 1 CUCCACCUUCGUGGGCGAGACAAACUGGCAAUUAGUUUAGUUUGUUUCUACUUAUGUGCAGUGUCCUUUGCUGUUGUUUAGCAAAUGACAGACUGA--------UAGACUGAUAGACUGA .(((((....))))).((((((((((((........))))))))))))......(((.(((((((.(((((((.....))))))).)))..)))))--------)).............. ( -30.90) >DroEre_CAF1 104276 104 - 1 CUCCACCUUCGUGGGCGAGACAAACUGGCAAUUAGUUUAGUUUGUUUCUACUUAUGUGCAGUGUCCUUUGCCGUUGUUUAGCAAAUGACAGAC----------------UGAUAGACUGA .(((((....))))).((((((((((((........))))))))))))......(((.(((((((.(((((.........))))).)))..))----------------)))))...... ( -26.90) >DroYak_CAF1 107569 120 - 1 CUCCACCUUCGUGGGCGAGACAAACUGGCAAUUAGUUUAGUUUGUUUCUACUUAUGUGCAGUGUCCUUUGCUGUUGUUUAGCAAAUGACAGACUGACGGACUGAUGGACUGAUAGACUGA .((((.((((((.(((((((((((((((........)))))))))))).............((((.(((((((.....))))))).))))).)).)))))..).))))............ ( -34.60) >consensus CUCCACCUUCGUGGGCGAGACAAACUGGCAAUUAGUUUAGUUUGUUUCUACUUAUGUGCAGUGUCCUUUGCUGUUGUUUAGCAAAUGACAGACUGA________U_GACUGAUAGACUGA ..((((....))))(((((((((((((((.....)).))))))))))).((....))))..((((.(((((((.....))))))).)))).............................. (-28.60 = -28.64 + 0.04)



| Location | 13,119,570 – 13,119,690 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -33.74 |
| Consensus MFE | -28.58 |
| Energy contribution | -28.50 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.653891 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13119570 120 + 23771897 UAAACAACAGCAAAGGACACUGCACAUAAGUAGAAACAAACUAUACUAAUUGCCAGUUUGUCUCGCCCACGAAGGUGAAGAACUUCUGGGGGCGAAGCGGCGCAGUCGUCGAACUCUUAG ...............((((((((.....((((...........))))....(((.....(..((((((..((((........))))...))))))..))))))))).))).......... ( -33.80) >DroSec_CAF1 107476 120 + 1 UAAACAACAGCAAAGGACACUGCACAUAAGUAGAAACAAACUAAACUAAUUGCCAGUUUGUCUCGCCCACGAAGGUGGAGAACUUCUGGGGCCGAAGCGGCGUAGUCGUCGAACUCUUAG ...............((((((((.((.((((.((.(((((((............))))))).))..((((....))))...)))).))..((((...))))))))).))).......... ( -32.30) >DroSim_CAF1 105630 120 + 1 UAAACAACAGCAAAGGACACUGCACAUAAGUAGAAACAAACUAAACUAAUUGCCAGUUUGUCUCGCCCACGAAGGUGGAGAACUUCUGGGGGCGAAGCGGCGUAGUCGUCGAACUCUUAG ...............((((((((.....(((........))).........(((..((((((((..((((....))))((.....)).))))))))..)))))))).))).......... ( -31.20) >DroEre_CAF1 104300 120 + 1 UAAACAACGGCAAAGGACACUGCACAUAAGUAGAAACAAACUAAACUAAUUGCCAGUUUGUCUCGCCCACGAAGGUGGAGAAGUUCCGGGGGCGAAGCGGCGUAGUCGUUGAACUCUAAG ....(((((((.....((.((((............(((((((............))))))).((((((((....))((((...))))..)))))).)))).)).)))))))......... ( -35.60) >DroYak_CAF1 107609 120 + 1 UAAACAACAGCAAAGGACACUGCACAUAAGUAGAAACAAACUAAACUAAUUGCCAGUUUGUCUCGCCCACGAAGGUGGAGAACUUCUGGGGGCGAGGCGGCGUAGUCGUCGAACUCUUAG ...............((((((((.....(((........))).........(((.....(((((((((..((((........))))...))))))))))))))))).))).......... ( -35.80) >consensus UAAACAACAGCAAAGGACACUGCACAUAAGUAGAAACAAACUAAACUAAUUGCCAGUUUGUCUCGCCCACGAAGGUGGAGAACUUCUGGGGGCGAAGCGGCGUAGUCGUCGAACUCUUAG ...............((((((((.....(((........))).........(((.(((....((((((..((((........))))...))))))))))))))))).))).......... (-28.58 = -28.50 + -0.08)


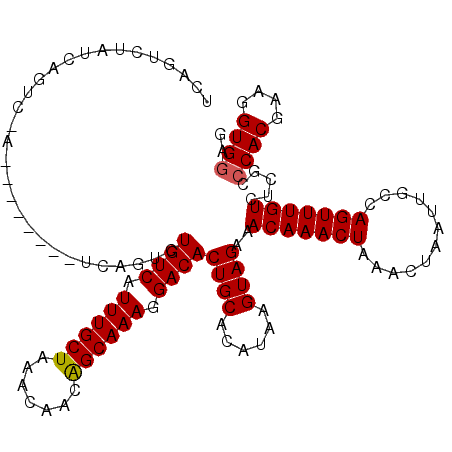
| Location | 13,119,570 – 13,119,690 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -35.32 |
| Consensus MFE | -30.00 |
| Energy contribution | -30.08 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.770242 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13119570 120 - 23771897 CUAAGAGUUCGACGACUGCGCCGCUUCGCCCCCAGAAGUUCUUCACCUUCGUGGGCGAGACAAACUGGCAAUUAGUAUAGUUUGUUUCUACUUAUGUGCAGUGUCCUUUGCUGUUGUUUA .(((.(((..(((.(((((((......((((...((((........))))..))))(((((((((((((.....)).))))))))))).......))))))))))....))).))).... ( -40.80) >DroSec_CAF1 107476 120 - 1 CUAAGAGUUCGACGACUACGCCGCUUCGGCCCCAGAAGUUCUCCACCUUCGUGGGCGAGACAAACUGGCAAUUAGUUUAGUUUGUUUCUACUUAUGUGCAGUGUCCUUUGCUGUUGUUUA .(((.(((..(((.(((.(((.((....))((((((((........)))).)))).((((((((((((........)))))))))))).......))).))))))....))).))).... ( -34.40) >DroSim_CAF1 105630 120 - 1 CUAAGAGUUCGACGACUACGCCGCUUCGCCCCCAGAAGUUCUCCACCUUCGUGGGCGAGACAAACUGGCAAUUAGUUUAGUUUGUUUCUACUUAUGUGCAGUGUCCUUUGCUGUUGUUUA .(((.(((..(((.(((.(((......((((...((((........))))..))))((((((((((((........)))))))))))).......))).))))))....))).))).... ( -34.20) >DroEre_CAF1 104300 120 - 1 CUUAGAGUUCAACGACUACGCCGCUUCGCCCCCGGAACUUCUCCACCUUCGUGGGCGAGACAAACUGGCAAUUAGUUUAGUUUGUUUCUACUUAUGUGCAGUGUCCUUUGCCGUUGUUUA .........(((((.(.((((.((...((((.((((...........)))).))))((((((((((((........)))))))))))).........)).)))).....).))))).... ( -32.50) >DroYak_CAF1 107609 120 - 1 CUAAGAGUUCGACGACUACGCCGCCUCGCCCCCAGAAGUUCUCCACCUUCGUGGGCGAGACAAACUGGCAAUUAGUUUAGUUUGUUUCUACUUAUGUGCAGUGUCCUUUGCUGUUGUUUA .(((.(((..(((.(((.(((...(((((((...((((........))))..)))))))(((((((((........)))))))))..........))).))))))....))).))).... ( -34.70) >consensus CUAAGAGUUCGACGACUACGCCGCUUCGCCCCCAGAAGUUCUCCACCUUCGUGGGCGAGACAAACUGGCAAUUAGUUUAGUUUGUUUCUACUUAUGUGCAGUGUCCUUUGCUGUUGUUUA ..........((((((.((((.((...((((...((((........))))..))))(((((((((((((.....)).))))))))))).........)).))))........)))))).. (-30.00 = -30.08 + 0.08)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:26:53 2006