
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,117,347 – 13,117,615 |
| Length | 268 |
| Max. P | 0.999912 |

| Location | 13,117,347 – 13,117,467 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.73 |
| Mean single sequence MFE | -24.30 |
| Consensus MFE | -24.14 |
| Energy contribution | -23.66 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928144 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13117347 120 + 23771897 AACGUUUAAACAUAUUGAAUUACUUUAAAAAAGAUCUUACCCCCCAACCCGGCGGCCAACGUCGCAAUUUGUUAAUUUCUUGUCAACACGAGUCGAAAGACUAAUUGGCCCGAAGAACCC ...((((.((((.((((.....(((.....)))................(((((.....))))))))).))))...(((..(((((....((((....))))..)))))..))))))).. ( -24.40) >DroSec_CAF1 105253 119 + 1 AACGUUUAAACAUAUUGAAUUACUUUAAAAAAGAUCUUACCC-CCAACCCGGCGGCCAACGUCGCAAUUUGUUAAUUUCUUGUCAACACGAGUCAAAAGACUAAUUGGCCCGAAGAACCC ...((((.((((.((((.....(((.....))).........-......(((((.....))))))))).))))...(((..(((((....((((....))))..)))))..))))))).. ( -20.70) >DroSim_CAF1 103359 119 + 1 AACGUUUAAACAUAUUGAAUUACUUUAAAAAAGAUCUUACCC-CCAACCCGGCGGCCAACGUCGCAAUUUGUUAAUUUCUUGUCAACACGAGUCGAAAGACUAAUUGGCCCGAAGAACCC ...((((.((((.((((.....(((.....))).........-......(((((.....))))))))).))))...(((..(((((....((((....))))..)))))..))))))).. ( -24.40) >DroEre_CAF1 101748 118 + 1 AGCGUUUAAACAUAUUGAAUUACUUUAAAAAAGAUAUUUGCC-CC-ACCCGGCGGCCAACGUCGCAAUUUGUUAAUUUCUUGUCAACACGAGUCGAAGGACUAAUUGGCCCGAGGAACCC .((((((((.....))))))............((..((.(((-((-....)).))).))..))))..........(((((((((((....((((....))))..)))))..))))))... ( -25.20) >DroYak_CAF1 105340 118 + 1 AACGUUUAAACAUAUUGAAUUACUUUAAAAGAGAUCUUUUCC-CC-ACCCGACGGCCAACGUCGCAAUUUGUUAAUUUCUUGUCAACACGAGUCGAAAGACUAAUUGGCCCGAAGAACCC ...((((.((((.((((.........(((((....)))))..-..-...(((((.....))))))))).))))...(((..(((((....((((....))))..)))))..))))))).. ( -26.80) >consensus AACGUUUAAACAUAUUGAAUUACUUUAAAAAAGAUCUUACCC_CCAACCCGGCGGCCAACGUCGCAAUUUGUUAAUUUCUUGUCAACACGAGUCGAAAGACUAAUUGGCCCGAAGAACCC ...((((.((((.((((.....(((.....)))................(((((.....))))))))).))))...(((..(((((....((((....))))..)))))..))))))).. (-24.14 = -23.66 + -0.48)


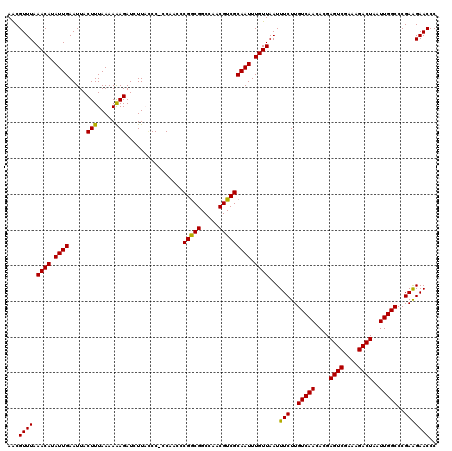
| Location | 13,117,427 – 13,117,547 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.00 |
| Mean single sequence MFE | -35.54 |
| Consensus MFE | -34.54 |
| Energy contribution | -34.30 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.29 |
| SVM RNA-class probability | 0.991783 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13117427 120 - 23771897 CGUUAAAUGAAAAUAUUCAGAGUGGCAAGAGGCUAAAUAUGGCCAAGUGAUUUAAAGGGUAAGAGUGACGGGGCCGAAGCGGGUUCUUCGGGCCAAUUAGUCUUUCGACUCGUGUUGACA .(((((.((((....))))((((.(..(((((((((...(((((..((.((((.........)))).))..))))).....(((.(....))))..)))))))))).))))...))))). ( -35.70) >DroSec_CAF1 105332 120 - 1 CGUUAAAUGAAAAUAUUCAGAGUGGCAAGAGGCUAAAUAUGGCCAAGUGAUUUAAAGGGUAAGAGUGACGGGGCCGAAGCGGGUUCUUCGGGCCAAUUAGUCUUUUGACUCGUGUUGACA .(((((.((((....))))((((..(((((((((((...(((((..((.((((.........)))).))..))))).....(((.(....))))..)))))))))))))))...))))). ( -37.70) >DroSim_CAF1 103438 120 - 1 CGUUAAAUGAAAAUAUUCAGAGUGGCAAGAGGCUAAAUAUGGCCAAGUGAUUUAAAGGGUAAGAGUGACGGGGCCGAAGCGGGUUCUUCGGGCCAAUUAGUCUUUCGACUCGUGUUGACA .(((((.((((....))))((((.(..(((((((((...(((((..((.((((.........)))).))..))))).....(((.(....))))..)))))))))).))))...))))). ( -35.70) >DroEre_CAF1 101826 120 - 1 CGUUAAAUGAAAAUAUUCAGAGUGGCAAGAGGCUAAAUAUGGCCAAGUGAUUUAAAGGGUAAGAGUGACGGCACUGAAGCGGGUUCCUCGGGCCAAUUAGUCCUUCGACUCGUGUUGACA .(((((.((((....))))((((.(.(((.((((((...(((((..((.((((.........)))).))((.(((......))).))...))))).)))))))))).))))...))))). ( -32.90) >DroYak_CAF1 105418 120 - 1 CGUUAAAUGAAAAUAUUCAGAGUGGCAAGAGGCUAAAUAUGGCCAAGUGAUUUAAAGGGUAAGAGUGACGGGGCCGAAGCGGGUUCUUCGGGCCAAUUAGUCUUUCGACUCGUGUUGACA .(((((.((((....))))((((.(..(((((((((...(((((..((.((((.........)))).))..))))).....(((.(....))))..)))))))))).))))...))))). ( -35.70) >consensus CGUUAAAUGAAAAUAUUCAGAGUGGCAAGAGGCUAAAUAUGGCCAAGUGAUUUAAAGGGUAAGAGUGACGGGGCCGAAGCGGGUUCUUCGGGCCAAUUAGUCUUUCGACUCGUGUUGACA .(((((.((((....))))((((.(..(((((((((...(((((..((.((((.........)))).))((((((......))))))...))))).)))))))))).))))...))))). (-34.54 = -34.30 + -0.24)

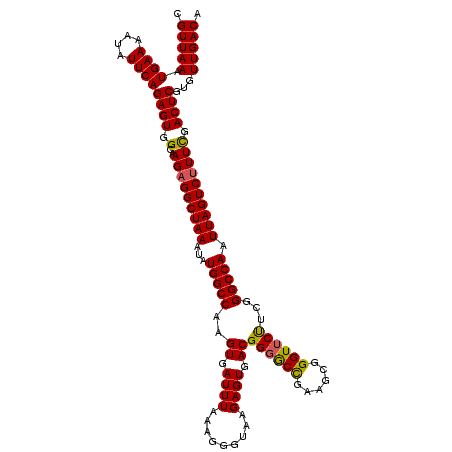
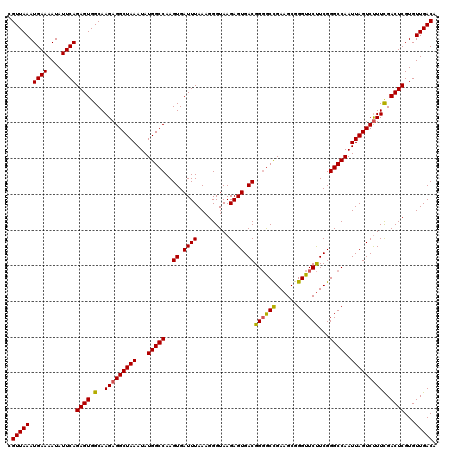
| Location | 13,117,467 – 13,117,575 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.36 |
| Mean single sequence MFE | -17.90 |
| Consensus MFE | -17.38 |
| Energy contribution | -17.06 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.97 |
| SVM decision value | 3.05 |
| SVM RNA-class probability | 0.998266 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13117467 108 - 23771897 AUGAUAAA------------AAACGAGGAAAAGUAAAUGACGUUAAAUGAAAAUAUUCAGAGUGGCAAGAGGCUAAAUAUGGCCAAGUGAUUUAAAGGGUAAGAGUGACGGGGCCGAAGC ........------------.............(((((.((((((..((((....))))...))))....(((((....)))))..)).)))))...(((..(.....)...)))..... ( -17.80) >DroSec_CAF1 105372 108 - 1 AUGAUAAA------------AAACGAGGAAAAGUAAAUGACGUUAAAUGAAAAUAUUCAGAGUGGCAAGAGGCUAAAUAUGGCCAAGUGAUUUAAAGGGUAAGAGUGACGGGGCCGAAGC ........------------.............(((((.((((((..((((....))))...))))....(((((....)))))..)).)))))...(((..(.....)...)))..... ( -17.80) >DroSim_CAF1 103478 108 - 1 AUGAUAAA------------AAACGAGGAAAAGUAAAUGACGUUAAAUGAAAAUAUUCAGAGUGGCAAGAGGCUAAAUAUGGCCAAGUGAUUUAAAGGGUAAGAGUGACGGGGCCGAAGC ........------------.............(((((.((((((..((((....))))...))))....(((((....)))))..)).)))))...(((..(.....)...)))..... ( -17.80) >DroEre_CAF1 101866 120 - 1 AUGAUAAAAAAAAAGAAAAAAAACGAGGAAAAGUAAAUGACGUUAAAUGAAAAUAUUCAGAGUGGCAAGAGGCUAAAUAUGGCCAAGUGAUUUAAAGGGUAAGAGUGACGGCACUGAAGC .................................(((((.((((((..((((....))))...))))....(((((....)))))..)).))))).........((((....))))..... ( -18.30) >DroYak_CAF1 105458 110 - 1 AUGAUAAAA----------AAAACGAGGAAAAGUAAAUGACGUUAAAUGAAAAUAUUCAGAGUGGCAAGAGGCUAAAUAUGGCCAAGUGAUUUAAAGGGUAAGAGUGACGGGGCCGAAGC .........----------..............(((((.((((((..((((....))))...))))....(((((....)))))..)).)))))...(((..(.....)...)))..... ( -17.80) >consensus AUGAUAAA____________AAACGAGGAAAAGUAAAUGACGUUAAAUGAAAAUAUUCAGAGUGGCAAGAGGCUAAAUAUGGCCAAGUGAUUUAAAGGGUAAGAGUGACGGGGCCGAAGC .................................(((((.((((((..((((....))))...))))....(((((....)))))..)).)))))...(((..(.....)...)))..... (-17.38 = -17.06 + -0.32)


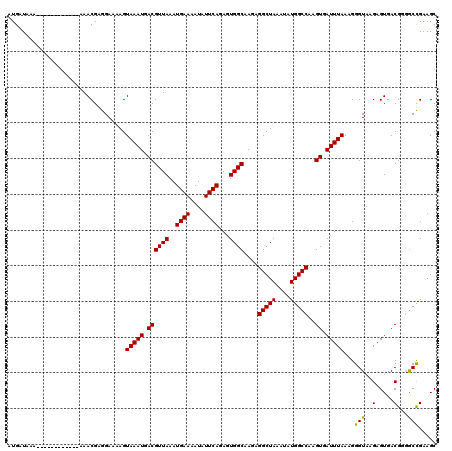
| Location | 13,117,507 – 13,117,615 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.77 |
| Mean single sequence MFE | -13.96 |
| Consensus MFE | -13.78 |
| Energy contribution | -13.62 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.99 |
| SVM decision value | 4.51 |
| SVM RNA-class probability | 0.999912 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13117507 108 + 23771897 AUAUUUAGCCUCUUGCCACUCUGAAUAUUUUCAUUUAACGUCAUUUACUUUUCCUCGUUU------------UUUAUCAUUCUUCAUUAAACUUUGUGGCGAAUGCUUGCUCCGCCGCAC ......(((...(((((((..((((....))))...((((...............)))).------------.......................)))))))..)))(((......))). ( -15.26) >DroSec_CAF1 105412 108 + 1 AUAUUUAGCCUCUUGCCACUCUGAAUAUUUUCAUUUAACGUCAUUUACUUUUCCUCGUUU------------UUUAUCAUUCUUCAUUAAACUUUGUGGCAAUUGCUUUCUCCACCGCAC ......(((...(((((((..((((....))))...((((...............)))).------------.......................)))))))..)))............. ( -13.86) >DroSim_CAF1 103518 108 + 1 AUAUUUAGCCUCUUGCCACUCUGAAUAUUUUCAUUUAACGUCAUUUACUUUUCCUCGUUU------------UUUAUCAUUCUUCAUUAAACUUUGUGGCGAAUGCUUUCUCCACCGCAC ......(((...(((((((..((((....))))...((((...............)))).------------.......................)))))))..)))............. ( -13.56) >DroEre_CAF1 101906 120 + 1 AUAUUUAGCCUCUUGCCACUCUGAAUAUUUUCAUUUAACGUCAUUUACUUUUCCUCGUUUUUUUUCUUUUUUUUUAUCAUUCUUCAUUAAACUUUGUGGCGAAUGCUUUCUCCACCGCAC ......(((...(((((((..((((....))))...((((...............))))....................................)))))))..)))............. ( -13.56) >DroYak_CAF1 105498 110 + 1 AUAUUUAGCCUCUUGCCACUCUGAAUAUUUUCAUUUAACGUCAUUUACUUUUCCUCGUUUU----------UUUUAUCAUUCUUCAUUAAACUUUGUGGCGAUUGCUUUCCCCACCGCCA ......(((...(((((((..((((....))))...((((...............))))..----------........................)))))))..)))............. ( -13.56) >consensus AUAUUUAGCCUCUUGCCACUCUGAAUAUUUUCAUUUAACGUCAUUUACUUUUCCUCGUUU____________UUUAUCAUUCUUCAUUAAACUUUGUGGCGAAUGCUUUCUCCACCGCAC ......(((...(((((((..((((....))))...((((...............))))....................................)))))))..)))............. (-13.78 = -13.62 + -0.16)

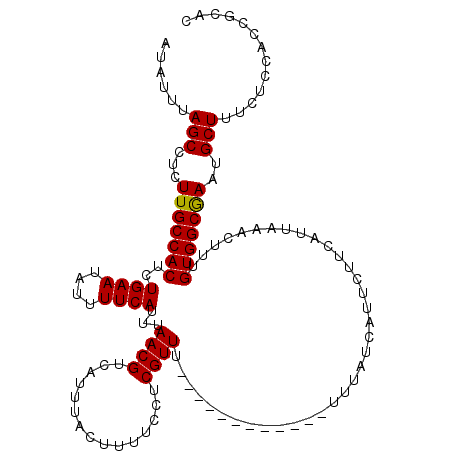
| Location | 13,117,507 – 13,117,615 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.77 |
| Mean single sequence MFE | -16.25 |
| Consensus MFE | -13.90 |
| Energy contribution | -13.90 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.685784 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13117507 108 - 23771897 GUGCGGCGGAGCAAGCAUUCGCCACAAAGUUUAAUGAAGAAUGAUAAA------------AAACGAGGAAAAGUAAAUGACGUUAAAUGAAAAUAUUCAGAGUGGCAAGAGGCUAAAUAU .(((......)))(((.((((((((...((((((((............------------....................))))))))(((....)))...)))))..))))))...... ( -17.35) >DroSec_CAF1 105412 108 - 1 GUGCGGUGGAGAAAGCAAUUGCCACAAAGUUUAAUGAAGAAUGAUAAA------------AAACGAGGAAAAGUAAAUGACGUUAAAUGAAAAUAUUCAGAGUGGCAAGAGGCUAAAUAU .............(((..(((((((...((((((((............------------....................))))))))(((....)))...)))))))...)))...... ( -18.25) >DroSim_CAF1 103518 108 - 1 GUGCGGUGGAGAAAGCAUUCGCCACAAAGUUUAAUGAAGAAUGAUAAA------------AAACGAGGAAAAGUAAAUGACGUUAAAUGAAAAUAUUCAGAGUGGCAAGAGGCUAAAUAU .............(((.((((((((...((((((((............------------....................))))))))(((....)))...)))))..))))))...... ( -14.55) >DroEre_CAF1 101906 120 - 1 GUGCGGUGGAGAAAGCAUUCGCCACAAAGUUUAAUGAAGAAUGAUAAAAAAAAAGAAAAAAAACGAGGAAAAGUAAAUGACGUUAAAUGAAAAUAUUCAGAGUGGCAAGAGGCUAAAUAU .((.((((((.......)))))).)).(((((..............................((........)).......((((..((((....))))...))))...)))))...... ( -14.30) >DroYak_CAF1 105498 110 - 1 UGGCGGUGGGGAAAGCAAUCGCCACAAAGUUUAAUGAAGAAUGAUAAAA----------AAAACGAGGAAAAGUAAAUGACGUUAAAUGAAAAUAUUCAGAGUGGCAAGAGGCUAAAUAU ((((.((((.((......)).))))........................----------......................((((..((((....))))...)))).....))))..... ( -16.80) >consensus GUGCGGUGGAGAAAGCAUUCGCCACAAAGUUUAAUGAAGAAUGAUAAA____________AAACGAGGAAAAGUAAAUGACGUUAAAUGAAAAUAUUCAGAGUGGCAAGAGGCUAAAUAU .............(((....(((((...((((((((............................................))))))))(((....)))...))))).....)))...... (-13.90 = -13.90 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:26:49 2006