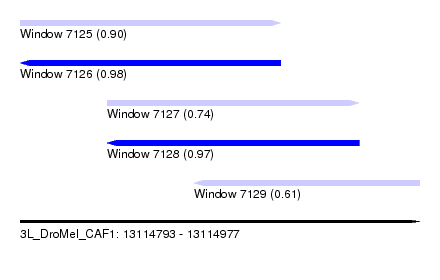
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,114,793 – 13,114,977 |
| Length | 184 |
| Max. P | 0.979069 |

| Location | 13,114,793 – 13,114,913 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.93 |
| Mean single sequence MFE | -39.22 |
| Consensus MFE | -34.37 |
| Energy contribution | -35.52 |
| Covariance contribution | 1.15 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.899258 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13114793 120 + 23771897 UGGCGGAGUCCAGUUAACGCUUCAAUGGUGGCAAAUGGCUAAAUUCGGGCGCAUAAAGUGAAUUGUUUUAUGCGCCGGGCGAAAUGCUUUCUCCGCUGCCUUUUUUCUUGGCGCCGAGUU .(((((((...(((...(((((......((((.....))))......(((((((((((.......))))))))))))))))....)))..)))))))(((.........)))........ ( -41.80) >DroSec_CAF1 102465 120 + 1 UGGCGGAGUCCAGUUAACGCUUCAAUGGUGGCAAAUGGCUAAAUUCGGGGGCAUAAAGUGAAUUGUUUUAUGCGCCGGGCGAAAUGCUUUCUCCGCUGCCUUUUUACUUGGCGCCGAGUU .(((((((...(((...(((((......((((.....))))......((.((((((((.......)))))))).)))))))....)))..)))))))........(((((....))))). ( -37.80) >DroSim_CAF1 100192 120 + 1 UUGCGGAGUCCAGUUAACGCUUCAAUGGUGGCAAAUGGCUAAAUUCGGGCGCAUAAAGUGAAUUGUUUUAUGCGCCGGGCGAAAUGCUUUCUCCGCUGCCUUUUUACUUGGCGCCGAGUU ..((((((...(((...(((((......((((.....))))......(((((((((((.......))))))))))))))))....)))..)))))).........(((((....))))). ( -40.60) >DroEre_CAF1 99088 96 + 1 UGGCGG-GUCCAGUUAACGCUUCAAUGGUGGCAAAUGGCUAAAUUCGGGCGCAUAAAGUGAAUUGUUUUAUGCGCCGGGCGAAAUGCUUUCUCCGCU----------------------- .(((((-(((((((((..(((........)))...))))).......(((((((((((.......)))))))))))))))(((.....))).)))))----------------------- ( -33.50) >DroYak_CAF1 102492 112 + 1 UGGCGG-GUCCAGUUAACGCUUCAAUGGUGGCAAAUGGCUAAAUUCGGGCGCAUAAAGUGAAUUGUUUUAUGCGCCCGGCGAAAUGCUUUCUCCGCUGCCU-------GGGCGCCAAGUU ((((..-((((((....((((.....))))......(((.......((((((((((((.......))))))))))))((.(((.....))).)))))..))-------)))))))).... ( -42.40) >consensus UGGCGGAGUCCAGUUAACGCUUCAAUGGUGGCAAAUGGCUAAAUUCGGGCGCAUAAAGUGAAUUGUUUUAUGCGCCGGGCGAAAUGCUUUCUCCGCUGCCUUUUU_CUUGGCGCCGAGUU .(((((((...(((...(((((......((((.....))))......(((((((((((.......))))))))))))))))....)))..)))))))(((.........)))........ (-34.37 = -35.52 + 1.15)


| Location | 13,114,793 – 13,114,913 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.93 |
| Mean single sequence MFE | -33.44 |
| Consensus MFE | -28.63 |
| Energy contribution | -29.38 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.979069 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13114793 120 - 23771897 AACUCGGCGCCAAGAAAAAAGGCAGCGGAGAAAGCAUUUCGCCCGGCGCAUAAAACAAUUCACUUUAUGCGCCCGAAUUUAGCCAUUUGCCACCAUUGAAGCGUUAACUGGACUCCGCCA .....((((((.........)))...((((...((..((((...((((((((((.........))))))))))))))....)).........(((((((....)))).))).))))))). ( -35.70) >DroSec_CAF1 102465 120 - 1 AACUCGGCGCCAAGUAAAAAGGCAGCGGAGAAAGCAUUUCGCCCGGCGCAUAAAACAAUUCACUUUAUGCCCCCGAAUUUAGCCAUUUGCCACCAUUGAAGCGUUAACUGGACUCCGCCA .....((((((.........)))...((((...((..((((...((.(((((((.........))))))))).))))....)).........(((((((....)))).))).))))))). ( -29.90) >DroSim_CAF1 100192 120 - 1 AACUCGGCGCCAAGUAAAAAGGCAGCGGAGAAAGCAUUUCGCCCGGCGCAUAAAACAAUUCACUUUAUGCGCCCGAAUUUAGCCAUUUGCCACCAUUGAAGCGUUAACUGGACUCCGCAA ........(((.........))).((((((...((..((((...((((((((((.........))))))))))))))....)).........(((((((....)))).))).)))))).. ( -35.30) >DroEre_CAF1 99088 96 - 1 -----------------------AGCGGAGAAAGCAUUUCGCCCGGCGCAUAAAACAAUUCACUUUAUGCGCCCGAAUUUAGCCAUUUGCCACCAUUGAAGCGUUAACUGGAC-CCGCCA -----------------------.((((.....((..((((...((((((((((.........))))))))))))))....)).........(((((((....)))).)))..-)))).. ( -25.90) >DroYak_CAF1 102492 112 - 1 AACUUGGCGCCC-------AGGCAGCGGAGAAAGCAUUUCGCCGGGCGCAUAAAACAAUUCACUUUAUGCGCCCGAAUUUAGCCAUUUGCCACCAUUGAAGCGUUAACUGGAC-CCGCCA ....(((((...-------.((((((((((......))))))((((((((((((.........))))))))))))............)))).(((((((....)))).)))..-.))))) ( -40.40) >consensus AACUCGGCGCCAAG_AAAAAGGCAGCGGAGAAAGCAUUUCGCCCGGCGCAUAAAACAAUUCACUUUAUGCGCCCGAAUUUAGCCAUUUGCCACCAUUGAAGCGUUAACUGGACUCCGCCA ........(((.........))).((((((...((..((((...((((((((((.........))))))))))))))....)).........(((((((....)))).))).)))))).. (-28.63 = -29.38 + 0.75)

| Location | 13,114,833 – 13,114,949 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.72 |
| Mean single sequence MFE | -33.24 |
| Consensus MFE | -24.57 |
| Energy contribution | -26.12 |
| Covariance contribution | 1.55 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.741081 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13114833 116 + 23771897 AAAUUCGGGCGCAUAAAGUGAAUUGUUUUAUGCGCCGGGCGAAAUGCUUUCUCCGCUGCCUUUUUUCUUGGCGCCGAGUUU----UCUUUUUUGAAAAACGAGUUACACACGUGUCGCAA .....(.(((((((((((.......))))))))))).)((((.(((....(((.((.(((.........))))).)))(((----((......)))))............))).)))).. ( -35.30) >DroSec_CAF1 102505 116 + 1 AAAUUCGGGGGCAUAAAGUGAAUUGUUUUAUGCGCCGGGCGAAAUGCUUUCUCCGCUGCCUUUUUACUUGGCGCCGAGUUU----UCUUUUUUGAAAAACGAGUUACACACGUGUCGCAA .....(.((.((((((((.......)))))))).)).)((((.(((....(((.((.(((.........))))).)))(((----((......)))))............))).)))).. ( -31.20) >DroSim_CAF1 100232 116 + 1 AAAUUCGGGCGCAUAAAGUGAAUUGUUUUAUGCGCCGGGCGAAAUGCUUUCUCCGCUGCCUUUUUACUUGGCGCCGAGUUU----UCUUUUUUGAAAAACGAGUUACACACGUGUCGCAA .....(.(((((((((((.......))))))))))).)((((.(((....(((.((.(((.........))))).)))(((----((......)))))............))).)))).. ( -35.30) >DroEre_CAF1 99127 94 + 1 AAAUUCGGGCGCAUAAAGUGAAUUGUUUUAUGCGCCGGGCGAAAUGCUUUCUCCGCU--------------------------UUCUUUUUUUGAAAAACGAGUUACACACGUGUCGCAA .....(.(((((((((((.......))))))))))).)((((.(((....(((...(--------------------------(((.......))))...))).......))).)))).. ( -26.40) >DroYak_CAF1 102531 113 + 1 AAAUUCGGGCGCAUAAAGUGAAUUGUUUUAUGCGCCCGGCGAAAUGCUUUCUCCGCUGCCU-------GGGCGCCAAGUUUUGUUUUUUUUUUGAAAAACGAGUUACACACGUGUCGCAA .....(((((((((((((.......)))))))))))))((((.(((........((.((..-------..)))).....((((((((((....)))))))))).......))).)))).. ( -38.00) >consensus AAAUUCGGGCGCAUAAAGUGAAUUGUUUUAUGCGCCGGGCGAAAUGCUUUCUCCGCUGCCUUUUU_CUUGGCGCCGAGUUU____UCUUUUUUGAAAAACGAGUUACACACGUGUCGCAA .....(.(((((((((((.......))))))))))).)((((........(((.((.(((.........))))).)))....................(((.(.....).))).)))).. (-24.57 = -26.12 + 1.55)



| Location | 13,114,833 – 13,114,949 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.72 |
| Mean single sequence MFE | -31.96 |
| Consensus MFE | -24.07 |
| Energy contribution | -25.30 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.966636 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13114833 116 - 23771897 UUGCGACACGUGUGUAACUCGUUUUUCAAAAAAGA----AAACUCGGCGCCAAGAAAAAAGGCAGCGGAGAAAGCAUUUCGCCCGGCGCAUAAAACAAUUCACUUUAUGCGCCCGAAUUU ..((((...((((.........(((((......))----)))(((.(((((.........))).)).)))...)))).))))..((((((((((.........))))))))))....... ( -33.50) >DroSec_CAF1 102505 116 - 1 UUGCGACACGUGUGUAACUCGUUUUUCAAAAAAGA----AAACUCGGCGCCAAGUAAAAAGGCAGCGGAGAAAGCAUUUCGCCCGGCGCAUAAAACAAUUCACUUUAUGCCCCCGAAUUU ..((((...((((.........(((((......))----)))(((.(((((.........))).)).)))...)))).)))).(((.(((((((.........)))))))..)))..... ( -27.90) >DroSim_CAF1 100232 116 - 1 UUGCGACACGUGUGUAACUCGUUUUUCAAAAAAGA----AAACUCGGCGCCAAGUAAAAAGGCAGCGGAGAAAGCAUUUCGCCCGGCGCAUAAAACAAUUCACUUUAUGCGCCCGAAUUU ..((((...((((.........(((((......))----)))(((.(((((.........))).)).)))...)))).))))..((((((((((.........))))))))))....... ( -33.50) >DroEre_CAF1 99127 94 - 1 UUGCGACACGUGUGUAACUCGUUUUUCAAAAAAAGAA--------------------------AGCGGAGAAAGCAUUUCGCCCGGCGCAUAAAACAAUUCACUUUAUGCGCCCGAAUUU ..((((...((((.....((((((((........)))--------------------------))))).....)))).))))..((((((((((.........))))))))))....... ( -26.80) >DroYak_CAF1 102531 113 - 1 UUGCGACACGUGUGUAACUCGUUUUUCAAAAAAAAAACAAAACUUGGCGCCC-------AGGCAGCGGAGAAAGCAUUUCGCCGGGCGCAUAAAACAAUUCACUUUAUGCGCCCGAAUUU ..((((...((((.......((((((......))))))....(((.((((..-------..)).)).)))...)))).))))((((((((((((.........))))))))))))..... ( -38.10) >consensus UUGCGACACGUGUGUAACUCGUUUUUCAAAAAAGA____AAACUCGGCGCCAAG_AAAAAGGCAGCGGAGAAAGCAUUUCGCCCGGCGCAUAAAACAAUUCACUUUAUGCGCCCGAAUUU .(((((.(((.(.....).)))...))...............(((.(((((.........))).)).)))...))).((((...((((((((((.........))))))))))))))... (-24.07 = -25.30 + 1.23)

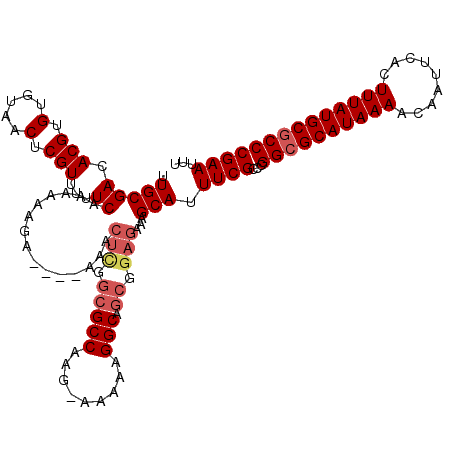

| Location | 13,114,873 – 13,114,977 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 85.21 |
| Mean single sequence MFE | -27.20 |
| Consensus MFE | -21.81 |
| Energy contribution | -23.04 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.610619 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13114873 104 - 23771897 UGGAUGGAUUGAGGGGCAACCCUACCAUUUGCGACACGUGUGUAACUCGUUUUUCAAAAAAGA----AAACUCGGCGCCAAGAAAAAAGGCAGCGGAGAAAGCAUUUC .((((((....((((....)))).))))))((...(((.(.....).)))(((((......))----)))(((.(((((.........))).)).)))...))..... ( -29.60) >DroSec_CAF1 102545 104 - 1 UGGAUGGAUUGAGGGGCAACCCUACCAUUUGCGACACGUGUGUAACUCGUUUUUCAAAAAAGA----AAACUCGGCGCCAAGUAAAAAGGCAGCGGAGAAAGCAUUUC .((((((....((((....)))).))))))((...(((.(.....).)))(((((......))----)))(((.(((((.........))).)).)))...))..... ( -29.60) >DroSim_CAF1 100272 104 - 1 UGGAUGGAUUGAGGGGCAACCCUACCAUUUGCGACACGUGUGUAACUCGUUUUUCAAAAAAGA----AAACUCGGCGCCAAGUAAAAAGGCAGCGGAGAAAGCAUUUC .((((((....((((....)))).))))))((...(((.(.....).)))(((((......))----)))(((.(((((.........))).)).)))...))..... ( -29.60) >DroEre_CAF1 99167 82 - 1 CGGAUGGAUUGAGGGGCAACCCUACCAUUUGCGACACGUGUGUAACUCGUUUUUCAAAAAAAGAA--------------------------AGCGGAGAAAGCAUUUC (((((((....((((....)))).)))))))......((((.....((((((((........)))--------------------------))))).....))))... ( -23.00) >DroYak_CAF1 102571 101 - 1 UGGAUGAAUUGAGGGGCAACCCUACCAUUUGCGACACGUGUGUAACUCGUUUUUCAAAAAAAAAACAAAACUUGGCGCCC-------AGGCAGCGGAGAAAGCAUUUC .(((((.....((((....))))..)))))((.(((....))).....((((((......))))))....(((.((((..-------..)).)).)))...))..... ( -24.20) >consensus UGGAUGGAUUGAGGGGCAACCCUACCAUUUGCGACACGUGUGUAACUCGUUUUUCAAAAAAGA____AAACUCGGCGCCAAG_AAAAAGGCAGCGGAGAAAGCAUUUC .((((((....((((....)))).))))))((((.(((.(.....).)))...))...............(((.(((((.........))).)).)))...))..... (-21.81 = -23.04 + 1.23)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:26:41 2006