| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
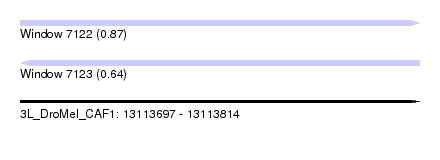
| Location | 13,113,697 – 13,113,814 |
| Length | 117 |
| Max. P | 0.870645 |

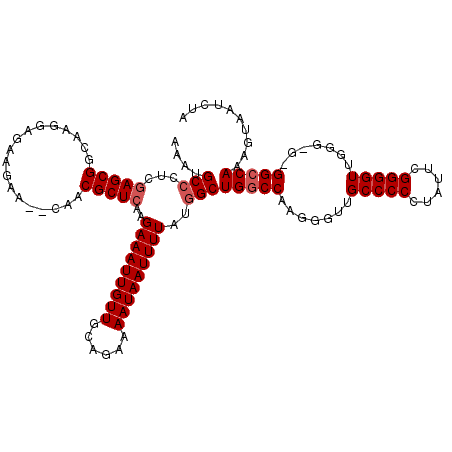
| Location | 13,113,697 – 13,113,814 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 95.03 |
| Mean single sequence MFE | -34.89 |
| Consensus MFE | -30.25 |
| Energy contribution | -30.85 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.870645 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
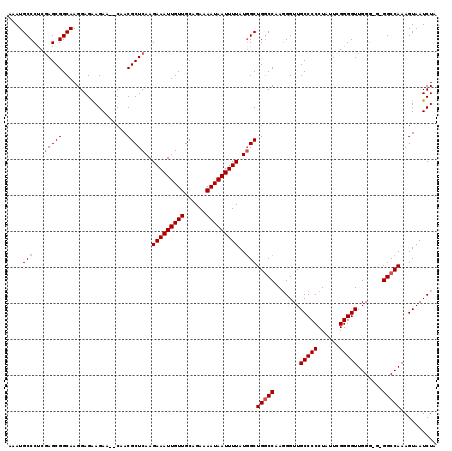
>3L_DroMel_CAF1 13113697 117 + 23771897 AAAUGCCCCCGAGCGGCAAGGAGAAGAA--CAACGCUCAAGAAAUUGUUGCAGAAAAUAAUUUUAUGGCUGGCCAAGGGUUGCCCCCUAUUCGGGGUUGGGGGUGGCCAAAGUAAUCUA ....(((.(((((((.............--...)))))..(((((((((......)))))))))..))..)))....(((..(((((...........)))))..)))........... ( -38.29) >DroSec_CAF1 101357 115 + 1 AAAUGCCCUCGAGCGGCAAGGAGAAGAA--CAACGCUCAAGAAAUUGUUGCAGAAAAUAAUUUUAUUGCUGGCCAAUGGUUGCCCCCUAUUCGGGGUUGGG-G-GGCCAAAGUAAUCUA ..........(((((.............--...)))))..(((((((((......)))))))))((((((((((...))))((((((((........))))-)-)))...))))))... ( -32.89) >DroSim_CAF1 99098 116 + 1 AAAUGCCCUCGAGCGGCAAGGAGAAGAA--CAACGCUCAAGAAAUUGUUGCAGAAAAUAAUUUUAUGGCUGGCCAAGGGUUGCCCCCUAUUCGGGGUUGGGGG-GGCCAAAGUAAUCUA ....(((((.(((((.............--...)))))..(((((((((......))))))))).(((....)))))))).(((((((..(....)...))))-)))............ ( -34.99) >DroEre_CAF1 97975 113 + 1 AAAUGCCCUCCAGCGGCAAGGAGAAGAA--CAACGCUCAAGAAAUUGUUGCAGAAAAUAAUUUUAUGGCUGUCCAAGGGUUGCCCCCUAUUCGGGGUUGG----GGCCAAAGUAAUCUA ....(((((...(((((..(((.(....--....((.((.(((((((((......))))))))).))))).)))....)))))((((.....))))..))----)))............ ( -30.20) >DroYak_CAF1 101372 116 + 1 AAAUGCCCCCGAGCGGCAAGGAGGAGAACACAACGCUCAAGAAAUUGUUGCAGAAAAUAAUUUUAUGGCUGGCCAAGGGUUGCCCCCUAUUUGGGGUUGGG---GGCCAAAGUAAUCUA ....(((((((((((((.....((....((....((.((.(((((((((......))))))))).)))))).))....)))))((((.....)))))))))---)))............ ( -38.10) >consensus AAAUGCCCUCGAGCGGCAAGGAGAAGAA__CAACGCUCAAGAAAUUGUUGCAGAAAAUAAUUUUAUGGCUGGCCAAGGGUUGCCCCCUAUUCGGGGUUGGG_G_GGCCAAAGUAAUCUA ....(((...(((((..................)))))..(((((((((......)))))))))..)))(((((.......(((((......))))).......))))).......... (-30.25 = -30.85 + 0.60)

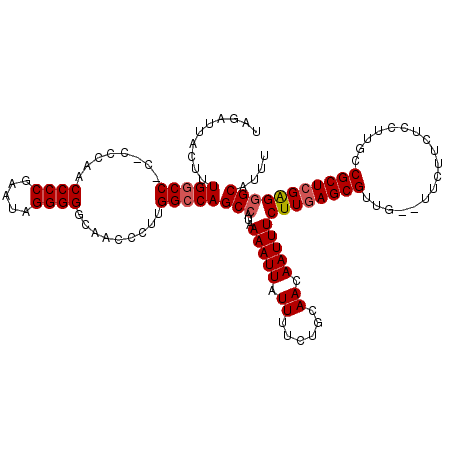
| Location | 13,113,697 – 13,113,814 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 95.03 |
| Mean single sequence MFE | -30.49 |
| Consensus MFE | -26.28 |
| Energy contribution | -26.64 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.638670 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13113697 117 - 23771897 UAGAUUACUUUGGCCACCCCCAACCCCGAAUAGGGGGCAACCCUUGGCCAGCCAUAAAAUUAUUUUCUGCAACAAUUUCUUGAGCGUUG--UUCUUCUCCUUGCCGCUCGGGGGCAUUU ..........((((((.......((((.....))))........))))))(((...(((((.((......)).)))))((((((((..(--..........)..))))))))))).... ( -31.36) >DroSec_CAF1 101357 115 - 1 UAGAUUACUUUGGCC-C-CCCAACCCCGAAUAGGGGGCAACCAUUGGCCAGCAAUAAAAUUAUUUUCUGCAACAAUUUCUUGAGCGUUG--UUCUUCUCCUUGCCGCUCGAGGGCAUUU ............(((-(-((............))))))........(((.(((..(((....)))..)))........((((((((..(--..........)..))))))))))).... ( -30.90) >DroSim_CAF1 99098 116 - 1 UAGAUUACUUUGGCC-CCCCCAACCCCGAAUAGGGGGCAACCCUUGGCCAGCCAUAAAAUUAUUUUCUGCAACAAUUUCUUGAGCGUUG--UUCUUCUCCUUGCCGCUCGAGGGCAUUU ..........(((((-.......((((.....)))).........)))))(((...(((((.((......)).)))))((((((((..(--..........)..))))))))))).... ( -30.19) >DroEre_CAF1 97975 113 - 1 UAGAUUACUUUGGCC----CCAACCCCGAAUAGGGGGCAACCCUUGGACAGCCAUAAAAUUAUUUUCUGCAACAAUUUCUUGAGCGUUG--UUCUUCUCCUUGCCGCUGGAGGGCAUUU .......(((..((.----.....(((.....)))(((((.....(((((((((..(((((.((......)).)))))..))...))))--)))......)))))))..)))....... ( -27.30) >DroYak_CAF1 101372 116 - 1 UAGAUUACUUUGGCC---CCCAACCCCAAAUAGGGGGCAACCCUUGGCCAGCCAUAAAAUUAUUUUCUGCAACAAUUUCUUGAGCGUUGUGUUCUCCUCCUUGCCGCUCGGGGGCAUUU ............(((---(((...(((.....)))(((((....(((....)))..............(((((..((....))..)))))..........)))))....)))))).... ( -32.70) >consensus UAGAUUACUUUGGCC_C_CCCAACCCCGAAUAGGGGGCAACCCUUGGCCAGCCAUAAAAUUAUUUUCUGCAACAAUUUCUUGAGCGUUG__UUCUUCUCCUUGCCGCUCGAGGGCAUUU ..........(((((........((((.....)))).........)))))(((...(((((.((......)).)))))((((((((..................))))))))))).... (-26.28 = -26.64 + 0.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:26:35 2006