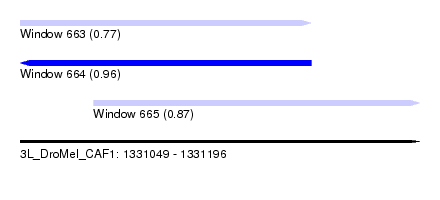
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,331,049 – 1,331,196 |
| Length | 147 |
| Max. P | 0.956167 |

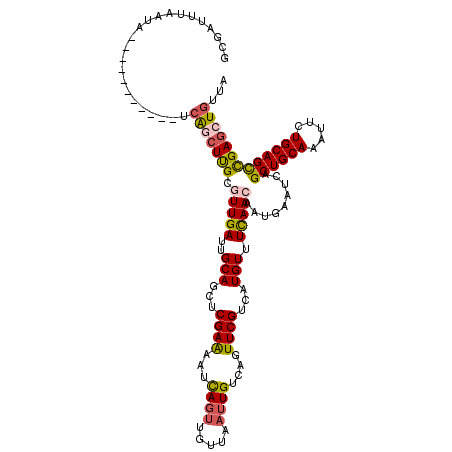
| Location | 1,331,049 – 1,331,156 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.65 |
| Mean single sequence MFE | -31.30 |
| Consensus MFE | -22.70 |
| Energy contribution | -23.18 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.766682 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1331049 107 + 23771897 --GGUUACCCA-----------UCAGCUCGCGUUGAUUGCAGCUCGAAAAUCAGUUGUUAAUUGUCAGUUCGUCAUGUUUCAACAAUGAAUCAGCUGCAAAUUCUGCAGCUGACCUGUUA --((....)).-----------.(((.....(((((..(((...((((...((((.....))))....))))...))).)))))......(((((((((.....))))))))).)))... ( -29.80) >DroSec_CAF1 3153 109 + 1 GCGAUUUAAUA-----------GCGGCUUGCGUUGAUUGCAGCUCGAAAAUCAGUUGUUAAUUGUCAGUUCGUCAUGUUUCAACCAUGAAUCAGCUGCAAAUUCUGCAGCCGAGAUGUUA ......(((((-----------.(((((.(((..(.((((((((((((...((((.....))))....))))(((((.......)))))...)))))))).)..))))))))...))))) ( -32.40) >DroSim_CAF1 3154 109 + 1 GCGAUUUAGUA-----------UCAGCUUGCGUUGAUUGCAGCUCGAAAAUUAGUUGUUAAUUGUCAGUUCGUCAUGUUUCAACCAUGAAUCAGCUGCAAAUUCUGCAGCCGAGAUGUUA ((((((.....-----------((((((.(((.....))))))).)).....)))))).....(((......(((((.......))))).((.((((((.....)))))).))))).... ( -25.90) >DroEre_CAF1 3133 116 + 1 GCGAUUUAAUAUGGUCAUCAGCUAAGCUUGCGUUGAUUGCAGCUCGAAAAUCAGUUG----UUGUCAGUUCGUCAUGUUUCAAAAAUGAAUCAGCUGCACGUUCUGCAGCCGAGCUGCUU ..((((......))))((((((.........)))))).((((((((....(((.(((----....((........))...)))...)))....((((((.....)))))))))))))).. ( -32.40) >DroYak_CAF1 3273 116 + 1 GCGAUUUAAUAUGGUUAGCCA-UCAGCUUGCGUUGAUUGCAGCUCGAGAAUCAGUUA---AAUGGCAGUUCGUCAUGUUUUAAAAAUGAAUCAGCUGCAAAUUAUGCAGUCGAGCUGCUA ...........(((....)))-(((((....)))))..(((((((((((.(((.(((---((((((.....)))))...))))...))).))..(((((.....)))))))))))))).. ( -36.00) >consensus GCGAUUUAAUA___________UCAGCUUGCGUUGAUUGCAGCUCGAAAAUCAGUUGUUAAUUGUCAGUUCGUCAUGUUUCAACAAUGAAUCAGCUGCAAAUUCUGCAGCCGAGCUGUUA .......................(((((((.(((((..(((...((((...((((.....))))....))))...))).))))).........((((((.....)))))))))))))... (-22.70 = -23.18 + 0.48)

| Location | 1,331,049 – 1,331,156 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.65 |
| Mean single sequence MFE | -29.89 |
| Consensus MFE | -20.11 |
| Energy contribution | -20.83 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.956167 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1331049 107 - 23771897 UAACAGGUCAGCUGCAGAAUUUGCAGCUGAUUCAUUGUUGAAACAUGACGAACUGACAAUUAACAACUGAUUUUCGAGCUGCAAUCAACGCGAGCUGA-----------UGGGUAACC-- ..((..(((((((((.....((((((((((.((((((((((..................))))))).)))...)).)))))))).....)).))))))-----------)..))....-- ( -32.47) >DroSec_CAF1 3153 109 - 1 UAACAUCUCGGCUGCAGAAUUUGCAGCUGAUUCAUGGUUGAAACAUGACGAACUGACAAUUAACAACUGAUUUUCGAGCUGCAAUCAACGCAAGCCGC-----------UAUUAAAUCGC ........(((((((.....((((((((...(((((.......))))).....(((.(((((.....))))).))))))))))).....)).))))).-----------........... ( -29.10) >DroSim_CAF1 3154 109 - 1 UAACAUCUCGGCUGCAGAAUUUGCAGCUGAUUCAUGGUUGAAACAUGACGAACUGACAAUUAACAACUAAUUUUCGAGCUGCAAUCAACGCAAGCUGA-----------UACUAAAUCGC .......((((((((.....((((((((...(((((.......))))).....(((.(((((.....))))).))))))))))).....)).))))))-----------........... ( -28.90) >DroEre_CAF1 3133 116 - 1 AAGCAGCUCGGCUGCAGAACGUGCAGCUGAUUCAUUUUUGAAACAUGACGAACUGACAA----CAACUGAUUUUCGAGCUGCAAUCAACGCAAGCUUAGCUGAUGACCAUAUUAAAUCGC ...(((((.((((((......(((((((..((((....))))......((((....((.----....))...)))))))))))......)).)))).))))).................. ( -29.90) >DroYak_CAF1 3273 116 - 1 UAGCAGCUCGACUGCAUAAUUUGCAGCUGAUUCAUUUUUAAAACAUGACGAACUGCCAUU---UAACUGAUUCUCGAGCUGCAAUCAACGCAAGCUGA-UGGCUAACCAUAUUAAAUCGC ..((((((((((((((.....)))))..((.(((.((......((........)).....---.)).))).))))))))))).((((.(....).)))-)((....))............ ( -29.10) >consensus UAACAGCUCGGCUGCAGAAUUUGCAGCUGAUUCAUUGUUGAAACAUGACGAACUGACAAUUAACAACUGAUUUUCGAGCUGCAAUCAACGCAAGCUGA___________UAUUAAAUCGC .......((((((((.....((((((((..((((....))))......((((....................)))))))))))).....))).)))))...................... (-20.11 = -20.83 + 0.72)

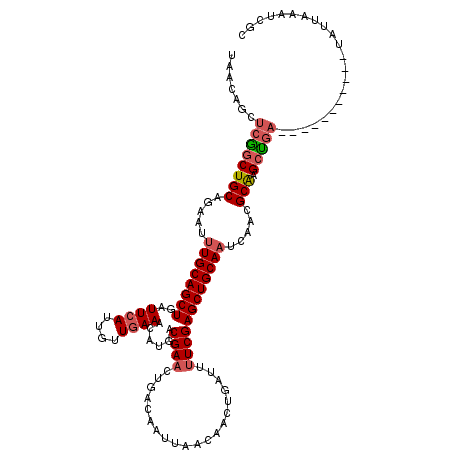
| Location | 1,331,076 – 1,331,196 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.39 |
| Mean single sequence MFE | -26.66 |
| Consensus MFE | -22.18 |
| Energy contribution | -23.14 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.873660 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1331076 120 + 23771897 AGCUCGAAAAUCAGUUGUUAAUUGUCAGUUCGUCAUGUUUCAACAAUGAAUCAGCUGCAAAUUCUGCAGCUGACCUGUUAUGAAAUAACUUUAAAGAGUCAUAUUUUUACGUAAUCUCCA .(((((((...((((.....))))....)))....(((((((..((((..(((((((((.....)))))))))..)))).)))))))........))))..................... ( -26.50) >DroSec_CAF1 3182 120 + 1 AGCUCGAAAAUCAGUUGUUAAUUGUCAGUUCGUCAUGUUUCAACCAUGAAUCAGCUGCAAAUUCUGCAGCCGAGAUGUUAUGAAAUAACUUCAAAGAGUCAUAUUCUUACGUAAUCUCCA .((((.......(((((((.....(((...((((....((((....))))((.((((((.....)))))).))))))...)))))))))).....))))..................... ( -24.20) >DroSim_CAF1 3183 120 + 1 AGCUCGAAAAUUAGUUGUUAAUUGUCAGUUCGUCAUGUUUCAACCAUGAAUCAGCUGCAAAUUCUGCAGCCGAGAUGUUAUGAAAUAACUUCAAAGAGUCAUAUUCUUACGUAAUCUCCA .((((.......(((((((.....(((...((((....((((....))))((.((((((.....)))))).))))))...)))))))))).....))))..................... ( -24.90) >DroEre_CAF1 3173 114 + 1 AGCUCGAAAAUCAGUUG----UUGUCAGUUCGUCAUGUUUCAAAAAUGAAUCAGCUGCACGUUCUGCAGCCGAGCUGCUUUUAAAUAACUUC--AGAGUCAUAUUCUUACGCAAUCUCCA .((((((.....(((((----((..(((((((...((.((((....)))).))((((((.....)))))))))))))......)))))))))--.))))..................... ( -26.40) >DroYak_CAF1 3312 117 + 1 AGCUCGAGAAUCAGUUA---AAUGGCAGUUCGUCAUGUUUUAAAAAUGAAUCAGCUGCAAAUUAUGCAGUCGAGCUGCUAUGAAAUAACAUUGAGGAGUCAUAAUCUUACGUAAUCUCCA .....((((....((((---.(((((((((((((((.........))))....((((((.....)))))))))))))))))....))))....((((.......))))......)))).. ( -31.30) >consensus AGCUCGAAAAUCAGUUGUUAAUUGUCAGUUCGUCAUGUUUCAACAAUGAAUCAGCUGCAAAUUCUGCAGCCGAGCUGUUAUGAAAUAACUUCAAAGAGUCAUAUUCUUACGUAAUCUCCA .((((.......(((((((......(((((((...((.((((....)))).))((((((.....)))))))))))))......))))))).....))))..................... (-22.18 = -23.14 + 0.96)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:42:15 2006