
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,105,428 – 13,105,539 |
| Length | 111 |
| Max. P | 0.800369 |

| Location | 13,105,428 – 13,105,539 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 85.92 |
| Mean single sequence MFE | -31.87 |
| Consensus MFE | -23.06 |
| Energy contribution | -23.88 |
| Covariance contribution | 0.82 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.42 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.800369 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13105428 111 + 23771897 UAACCAAGUUUAUGGUCAUAGAAACUUAGCACCGGCGACCCCGGAAUCCCA--------GCAUCCCGGAAAGGAAAGAGCAAAGUUCCUGGCUAAGUUGACCAUAAAAUUGCACACAAA ....(((.((((((((((.....(((((((.((((.((..(.((....)).--------)..))))))..(((((.........))))).))))))))))))))))).)))........ ( -35.20) >DroSec_CAF1 93094 111 + 1 UAACCAAGUUUAUGGUCAUAGAAACUUAGCACCGGCGACCCCAGAACCCCA--------GCAUCCCGGAUAGGAAAGAGCAAAGUUCCUGUCUAAGUUGACCAUAAAAUUGCACACAAA ....(((.((((((((((.....((((((..((((.((..(..(....)..--------)..))))))(((((((.........))))))))))))))))))))))).)))........ ( -28.50) >DroSim_CAF1 90863 111 + 1 UAACCAAGUUUAUGGUCAUAGAAACUUAGCACCGGCGACCCCGGAACCCCA--------GCAUCCCGGAUAGGAAAGAGCAAAGUUCCUGGCUAAGUUGACCAUAAAAUUGCACACAAA ....(((.((((((((((.....(((((((.((((.((..(.((....)).--------)..)))))).((((((.........))))))))))))))))))))))).)))........ ( -36.30) >DroEre_CAF1 89456 119 + 1 UAACCAAGUUUAUGGUCAUAGAAACUUAGCACCGGCGACCCCGGAACCCCGGAACGACGGCAUCCCGGAAAGGAAAGAGCAAAGUUCCUGGCCAAGUUGACCAUAAAAUUGCACACAAA ....(((.((((((((((.....((((.((.((((.....))))......(((((....((.((((.....))...))))...)))))..)).)))))))))))))).)))........ ( -34.20) >DroYak_CAF1 92758 111 + 1 UAACCAAGUUUAUGGUCAUAGAAACUUAGCACCGGCGACCCCGGAACCCCA--------GCAUCCCGGAAAGGAAAGAGCAAAGUUCCUGGCUAAGUUGACCAUAAAAUUGCACACAAA ....(((.((((((((((.....(((((((.((((.((..(.((....)).--------)..))))))..(((((.........))))).))))))))))))))))).)))........ ( -35.20) >DroAna_CAF1 86007 91 + 1 UAACCAAGUUUAUGGUCAUAGAAACUUAACCCAC-------------------------ACACACC---AAGGAUAUGGGCAAGUUGCAGGCUAAGUUGACCAUAAAAUUGCACACAAA ....(((.((((((((((.....(((((.((...-------------------------.....((---(......)))((.....)).)).))))))))))))))).)))........ ( -21.80) >consensus UAACCAAGUUUAUGGUCAUAGAAACUUAGCACCGGCGACCCCGGAACCCCA________GCAUCCCGGAAAGGAAAGAGCAAAGUUCCUGGCUAAGUUGACCAUAAAAUUGCACACAAA ....(((.((((((((((.....(((((((.((((.....))))...............((.((((.....))...))))..........))))))))))))))))).)))........ (-23.06 = -23.88 + 0.82)

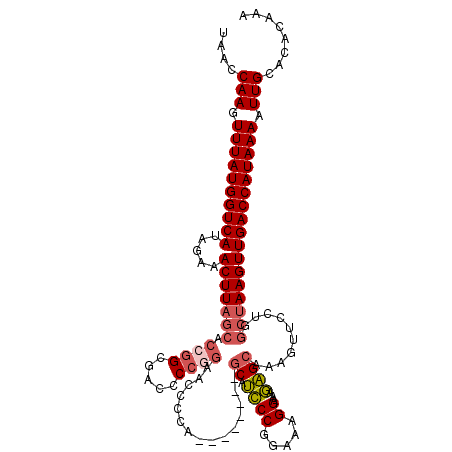

| Location | 13,105,428 – 13,105,539 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 85.92 |
| Mean single sequence MFE | -39.28 |
| Consensus MFE | -27.56 |
| Energy contribution | -28.85 |
| Covariance contribution | 1.29 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.56 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.779415 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13105428 111 - 23771897 UUUGUGUGCAAUUUUAUGGUCAACUUAGCCAGGAACUUUGCUCUUUCCUUUCCGGGAUGC--------UGGGAUUCCGGGGUCGCCGGUGCUAAGUUUCUAUGACCAUAAACUUGGUUA ........(((.(((((((((((((((((.(((((.........)))))..(((((((.(--------(((....)))).))).)))).))))))).....)))))))))).))).... ( -41.10) >DroSec_CAF1 93094 111 - 1 UUUGUGUGCAAUUUUAUGGUCAACUUAGACAGGAACUUUGCUCUUUCCUAUCCGGGAUGC--------UGGGGUUCUGGGGUCGCCGGUGCUAAGUUUCUAUGACCAUAAACUUGGUUA ........(((.((((((((((((((((.((((((.........)))))..(((((((.(--------..(....)..).))).)))).))))))).....)))))))))).))).... ( -36.20) >DroSim_CAF1 90863 111 - 1 UUUGUGUGCAAUUUUAUGGUCAACUUAGCCAGGAACUUUGCUCUUUCCUAUCCGGGAUGC--------UGGGGUUCCGGGGUCGCCGGUGCUAAGUUUCUAUGACCAUAAACUUGGUUA ........(((.(((((((((((((((((.(((((.........)))))..(((((((.(--------(((....)))).))).)))).))))))).....)))))))))).))).... ( -41.70) >DroEre_CAF1 89456 119 - 1 UUUGUGUGCAAUUUUAUGGUCAACUUGGCCAGGAACUUUGCUCUUUCCUUUCCGGGAUGCCGUCGUUCCGGGGUUCCGGGGUCGCCGGUGCUAAGUUUCUAUGACCAUAAACUUGGUUA ........(((.(((((((((((((((((((((((.........)))))(((((((((.(((......))).)))))))))......).))))))).....)))))))))).))).... ( -42.60) >DroYak_CAF1 92758 111 - 1 UUUGUGUGCAAUUUUAUGGUCAACUUAGCCAGGAACUUUGCUCUUUCCUUUCCGGGAUGC--------UGGGGUUCCGGGGUCGCCGGUGCUAAGUUUCUAUGACCAUAAACUUGGUUA ........(((.(((((((((((((((((.(((((.........)))))..(((((((.(--------(((....)))).))).)))).))))))).....)))))))))).))).... ( -41.10) >DroAna_CAF1 86007 91 - 1 UUUGUGUGCAAUUUUAUGGUCAACUUAGCCUGCAACUUGCCCAUAUCCUU---GGUGUGU-------------------------GUGGGUUAAGUUUCUAUGACCAUAAACUUGGUUA ........(((.((((((((((((((((((..((....(((((......)---)).)).)-------------------------)..)))))))).....)))))))))).))).... ( -33.00) >consensus UUUGUGUGCAAUUUUAUGGUCAACUUAGCCAGGAACUUUGCUCUUUCCUUUCCGGGAUGC________UGGGGUUCCGGGGUCGCCGGUGCUAAGUUUCUAUGACCAUAAACUUGGUUA ........(((.(((((((((((((((((.(((((.........))))).((((((((.(..........).)))))))).........))))))).....)))))))))).))).... (-27.56 = -28.85 + 1.29)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:26:26 2006