
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,091,419 – 13,091,556 |
| Length | 137 |
| Max. P | 0.711197 |

| Location | 13,091,419 – 13,091,525 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.90 |
| Mean single sequence MFE | -32.60 |
| Consensus MFE | -21.30 |
| Energy contribution | -21.22 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.711197 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
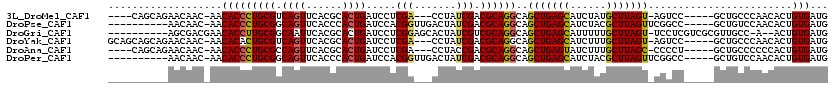
>3L_DroMel_CAF1 13091419 106 + 23771897 ----CAGCAGAACAAC-AACACCCUGCGUCAGUUCACGCACUGAUCCUCGA---CCUAUCGACGCAGGCAGCUGAGCAUCUAUGCUUAGU-AGUCC-----GCUGCCCAACACUGUGAUG ----((((.((.....-.....(((((((((((......)))))...((((---....))))))))))..((((((((....))))))))-..)).-----))))............... ( -31.70) >DroPse_CAF1 68609 104 + 1 ----------AACAAC-AACACCCUGCGGCAGUUCACCCACUGAUCCACGGUUGACUAUCGACGCAGGCAGCUGAGCAUCUACGCUUAGUUCGGCC-----GCUGUCCAACACUGUGAUG ----------......-..(((..((.(..((((.(((...........))).))))..)(((((.(((.(((((((......)))))))...)))-----)).)))...))..)))... ( -28.00) >DroGri_CAF1 110258 106 + 1 ----------AGCGACGAACACCUUGCGGCAAUUCACGCACUGAUCCUCGGAGCACUAUCGUCGCAGGCAGCUGAGCAUUUUUGCUUAGU-UCCUCGUCGCGUUGCC-A--ACUGUGAUG ----------.(((((((......((((((.((....((.(((.....))).))...)).))))))((.(((((((((....))))))))-))))))))))((..(.-.--...)..)). ( -40.50) >DroYak_CAF1 78118 110 + 1 GCAGCAGCAGAACAAC-AACACACUGCGUCAGUUCACGCACUGAUCCUCGA---CCUAUCGACGCAGGCAGCUGAGCAUCUUUGCUUAGU-AGUCC-----GCUGCCCAACACUGUGAUG ...(((((........-......((((((((((......)))))...((((---....)))))))))((.((((((((....))))))))-.))..-----))))).............. ( -34.70) >DroAna_CAF1 72834 106 + 1 ----CAGCAGAACAAC-AACACCCUGCGCCAGUUCACGCACUGAUCCUCGA---CCUACCGACGCAGGCAGCUGAGUAUCUUUGCUUAGC-CCCCU-----GCUGCCCCCCACUGUGAUG ----((((((......-.....((((((.((((......))))....(((.---.....)))))))))..((((((((....))))))))-...))-----))))............... ( -32.70) >DroPer_CAF1 44071 104 + 1 ----------AACAAC-AACACCCUGCGGCAGUUCACCCACUGAUCCACGGUUGACUAUCGACGCAGGCAGCUGAGCAUCUACGCUUAGUUCGGCC-----GCUGUCCAACACUGUGAUG ----------......-..(((..((.(..((((.(((...........))).))))..)(((((.(((.(((((((......)))))))...)))-----)).)))...))..)))... ( -28.00) >consensus __________AACAAC_AACACCCUGCGGCAGUUCACGCACUGAUCCUCGA___ACUAUCGACGCAGGCAGCUGAGCAUCUAUGCUUAGU_CGCCC_____GCUGCCCAACACUGUGAUG ...................(((((((((.((((......)))).....(((.......))).))))))..(((((((......)))))))........................)))... (-21.30 = -21.22 + -0.08)


| Location | 13,091,454 – 13,091,556 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 78.95 |
| Mean single sequence MFE | -22.44 |
| Consensus MFE | -15.88 |
| Energy contribution | -15.60 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.02 |
| Structure conservation index | 0.71 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.507202 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13091454 102 + 23771897 CUGAUCCUCGA---CCUAUCGACGCAGGCAGCUGAGCAUCUAUGCUUAGUAGUCC-----GCUGCCCAACACUGUGAUGCCAAAACCCCC-------AAGAAGCCCACUAAAACCAC-- .......((((---....)))).((.(((.((((((((....)))))))).))).-----))...........(((..((..........-------.....)).))).........-- ( -19.16) >DroGri_CAF1 110288 109 + 1 CUGAUCCUCGGAGCACUAUCGUCGCAGGCAGCUGAGCAUUUUUGCUUAGUUCCUCGUCGCGUUGCC-A--ACUGUGAUGCCAAAUUCUGU-------AAAUGCCCCACAACAACAAGAA .........((.(((.....(.((.(((.(((((((((....)))))))))))))).)((((..(.-.--...)..))))..........-------...))).))............. ( -30.90) >DroSim_CAF1 76723 102 + 1 CUGAUCCUCGA---CCUAUCGACGCAGGCAGCUGAGCAUCUAUGCUUAGUAGUCC-----GCUGCCCAACACUGUGAUGCCAAAACCCCC-------AAAAAGCCCACUAAAACCAC-- .......((((---....)))).((.(((.((((((((....)))))))).))).-----))...........(((..((..........-------.....)).))).........-- ( -19.16) >DroEre_CAF1 75680 102 + 1 CUGAUCCUCGA---CCUAUAGACGCAGGCAGCUGAGCAUCUAUGCUUAGUAGUCC-----GCUGCCCAACACUGUGAUGCCAAAACCCCC-------AAAAAACACACUAAAACCAC-- ...........---....(((..((.(((.((((((((....)))))))).))).-----))..........((((..............-------......))))))).......-- ( -17.95) >DroYak_CAF1 78157 102 + 1 CUGAUCCUCGA---CCUAUCGACGCAGGCAGCUGAGCAUCUUUGCUUAGUAGUCC-----GCUGCCCAACACUGUGAUGCCAAAACCCCC-------AAAAAGCCCACUAAAACCAC-- .......((((---....)))).((.(((.((((((((....)))))))).))).-----))...........(((..((..........-------.....)).))).........-- ( -19.96) >DroAna_CAF1 72869 109 + 1 CUGAUCCUCGA---CCUACCGACGCAGGCAGCUGAGUAUCUUUGCUUAGCCCCCU-----GCUGCCCCCCACUGUGAUGCCUGGUCCUGCCCCACCCUCACAUCCUGCCCCAUCCAC-- .......(((.---.....))).(((((..((((((((....))))))))..)))-----))..........(((((.(..(((.......)))..))))))...............-- ( -27.50) >consensus CUGAUCCUCGA___CCUAUCGACGCAGGCAGCUGAGCAUCUAUGCUUAGUAGUCC_____GCUGCCCAACACUGUGAUGCCAAAACCCCC_______AAAAAGCCCACUAAAACCAC__ ......................((((((((((((((((....))))))............))))))......))))........................................... (-15.88 = -15.60 + -0.28)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:26:16 2006