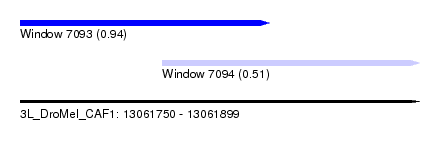
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,061,750 – 13,061,899 |
| Length | 149 |
| Max. P | 0.939068 |

| Location | 13,061,750 – 13,061,843 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 77.02 |
| Mean single sequence MFE | -25.44 |
| Consensus MFE | -11.68 |
| Energy contribution | -12.68 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.46 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939068 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
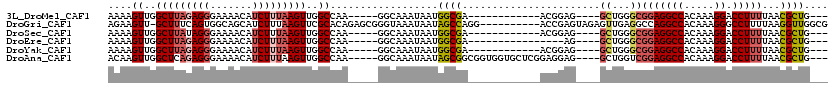
>3L_DroMel_CAF1 13061750 93 + 23771897 UUCCAUUUUCCUUUUUGCUAAGCAAAUAAUUGAGAGCGAGAC----UGG-GGGUGGUGAAAAGUUGGCUUAGAGGGAAAACAUCUUUAAGUUGGCCAA ..(((((..(..(((((((...(((....)))..))))))).----.).-.)))))......((..(((((((((.......)))))))))..))... ( -30.40) >DroGri_CAF1 70625 74 + 1 AUAAAUUCUUCUACUUGCUGAGCAAAUAACUGUAC-----------------------AGAAGUU-GCUUUCAGUGGCAGCAUCUUUAAGUUCGCACA ...............(((.((((......((....-----------------------))..(((-(((......))))))........))))))).. ( -14.40) >DroSec_CAF1 49416 93 + 1 UUCCAUUUUCCUUUUUGCUAAGCAAAUAAUUGAGAGCGAGAC----UGG-GGGUGGUGAAAAGUUGGCUUAUAGGGAAAACAUCUUUAAGUUGGCCAA ..(((((..(..(((((((...(((....)))..))))))).----.).-.)))))......((..(((((.(((.......))).)))))..))... ( -27.00) >DroEre_CAF1 46055 94 + 1 UUCCAUUUUCCUUUUUGCUAAGCAAAUAAUUGGGAGGGGGCC----UGGGUGGUGGUGAAAAGUUGGCUUAGAGGGAAAACAUCUUUAAGUUGGCCAA ..((((..(((..(((((...))))).....(((......))----))))..))))......((..(((((((((.......)))))))))..))... ( -25.50) >DroYak_CAF1 47931 98 + 1 UUCCAUUUUCCUUUUUGCUAAGCAAAUAACUGAAUGGGGGAAACGGGGGGGGGUGGUGAAAAGUUGGCUUAGAGGGAAAACAUCUUUAAGUUGGCCAA ..(((((..(((((((((...)))))............(....).))))..)))))......((..(((((((((.......)))))))))..))... ( -29.90) >consensus UUCCAUUUUCCUUUUUGCUAAGCAAAUAAUUGAGAGCGAGAC____UGG_GGGUGGUGAAAAGUUGGCUUAGAGGGAAAACAUCUUUAAGUUGGCCAA .............(((((...)))))....................................((..(((((((((.......)))))))))..))... (-11.68 = -12.68 + 1.00)


| Location | 13,061,803 – 13,061,899 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.19 |
| Mean single sequence MFE | -28.23 |
| Consensus MFE | -18.83 |
| Energy contribution | -19.58 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.67 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.512852 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13061803 96 + 23771897 AAAAGUUGGCUUAGAGGGAAAACAUCUUUAAGUUGGCCAA-----GGCAAAUAAUGGCGA------------ACGGAG----GCUGGGCGGAGGCCACAAAGGACCUUUUAACGCUG--- ....((..(((((((((.......)))))))))..))...-----..........((((.------------..((((----(((.(((....))).....)).)))))...)))).--- ( -28.40) >DroGri_CAF1 70660 109 + 1 AGAAGUU-GCUUUCAGUGGCAGCAUCUUUAAGUUCGCACAGAGCGGGUAAAUAAUAGCCAGG----------ACCGAGUAGAGUUGAGGCCAGGCCACAAAGGGCCUUUUAAGGUUGGCG (((.(((-(((......)))))).)))....(..(((.....)))..)........((((((----------.((..(......)..))))(((((......)))))........)))). ( -32.80) >DroSec_CAF1 49469 96 + 1 AAAAGUUGGCUUAUAGGGAAAACAUCUUUAAGUUGGCCAA-----GGCAAAUAAUGGCGA------------ACGGAG----GCUGGGCGGAGGCCACAAAGGACCUUUUAACGCUG--- .....(..((((.((((((.....)))))).(((.((((.-----.........)))).)------------))..))----))..)(((((((((.....)).))))....)))..--- ( -25.30) >DroEre_CAF1 46109 92 + 1 AAAAGUUGGCUUAGAGGGAAAACAUCUUUAAGUUGGCCAA-----GGCAAAUAAUGGCGA----------------AG----GCUGGGCGGAGGCCACAAAGGACCUUUUAACGCUG--- ....((..(((((((((.......)))))))))..))...-----..........(((..----------------..----))).((((((((((.....)).))))....)))).--- ( -28.00) >DroYak_CAF1 47989 96 + 1 AAAAGUUGGCUUAGAGGGAAAACAUCUUUAAGUUGGCCAA-----GGCAAAUAAUGGCGA------------ACGGAG----GCUGGGCGGAGGCCACAAAGGACCUUUUAACGCUG--- ....((..(((((((((.......)))))))))..))...-----..........((((.------------..((((----(((.(((....))).....)).)))))...)))).--- ( -28.40) >DroAna_CAF1 43441 108 + 1 ACAAGUUGGCUCAGAGGGAAAACAUCUUUAAGUUGGCCAA-----GGCAAAUAAUAGCGGCGGUGGUGCUCGGAGGAG----GCUGGUCGGAGGCCACAAAGGACCUUUUAACGCUG--- ....((..(((.(((((.......))))).)))..))...-----............((((((((((.(((.((....----.....)).))))))))((((....))))..)))))--- ( -26.50) >consensus AAAAGUUGGCUUAGAGGGAAAACAUCUUUAAGUUGGCCAA_____GGCAAAUAAUGGCGA____________ACGGAG____GCUGGGCGGAGGCCACAAAGGACCUUUUAACGCUG___ ....((..(((((((((.......)))))))))..))..................((((.......................((...))(((((((.....)).)))))...)))).... (-18.83 = -19.58 + 0.75)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:26:06 2006