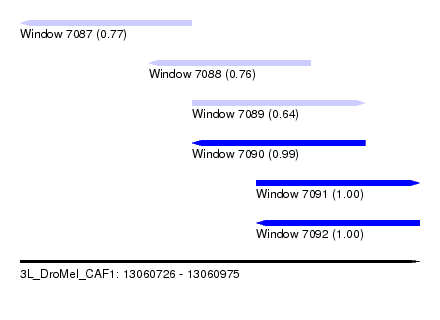
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,060,726 – 13,060,975 |
| Length | 249 |
| Max. P | 0.999925 |

| Location | 13,060,726 – 13,060,833 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 93.74 |
| Mean single sequence MFE | -34.08 |
| Consensus MFE | -28.56 |
| Energy contribution | -28.84 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.766595 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13060726 107 - 23771897 UUAAGCACGAGCACAGCGGCCUAUCCUUGGGCAUCAUGGAAGAUGGCUACUACAGCAGUGGAAUCACCAUCGGUAUCCUGACCACAUUUGCUGUGAUUAGGCUUCUU ..((((.....((((((((((((....))))).(((.(((.((((((((((.....))))).....)))))....)))))).......))))))).....))))... ( -35.00) >DroSec_CAF1 48400 107 - 1 UUAAGCACGAGCACAGCGGCCUAUCCUUGGGCAUCAUGGAGGACGGCUACUACAGCAGUGGAAUCACCAUCGGUAUCCUGACCACAUUUGCUGUGAUUAGGCUUCUU ..((((.....((((((((((((....))))).(((.(((...(((((.....))).((((.....))))))...)))))).......))))))).....))))... ( -33.70) >DroSim_CAF1 44194 107 - 1 UUAAGCACGAGCACAGCGGCCUAUCCUUGGGCAUCAUGGAGGAUGGCUACUAUAGCAGUGGAAUCACCAUCGGUAUCCUGACCACAUUUGCUGUGAUUAGGCUUCUU ..((((.....((((((((((((....))))).(((.(((.((((((((((.....))))).....)))))....)))))).......))))))).....))))... ( -35.00) >DroEre_CAF1 45092 107 - 1 UGAAGCACGAGCACAGCGGCUUAUCCUUGGGCAUCUUGGAGGAUGGUUACUACAGCAGUGGAAUCACCAUCGGUAUCCUGACCACAUUUGCUGUGAUAAGGUUUCUG .(((((.....((((((((((((....)))))....(((.((.(((((.((((....))))))))))).((((....)))))))....))))))).....))))).. ( -34.20) >DroYak_CAF1 46967 107 - 1 UCAAGCACGAGCACAGCGGCUUAUCCUUGGGCAUCAUGGAGGAUGGCUACUACAGCAGUGGAAUCACCAUUGGCAUACUGACCACAUUUGCUGUGAUAAGACUGCUU ....((....))..(((((((((((..(((.((((......)))).)))..((((((((((.....))))((((.....).)))....)))))))))))).))))). ( -32.50) >consensus UUAAGCACGAGCACAGCGGCCUAUCCUUGGGCAUCAUGGAGGAUGGCUACUACAGCAGUGGAAUCACCAUCGGUAUCCUGACCACAUUUGCUGUGAUUAGGCUUCUU ..((((.....((((((((((((....)))))....((((((((((((.....))).((((.....))))...))))))..)))....))))))).....))))... (-28.56 = -28.84 + 0.28)

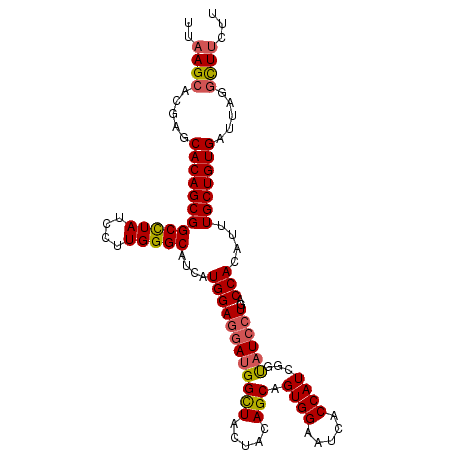

| Location | 13,060,806 – 13,060,907 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 92.77 |
| Mean single sequence MFE | -36.92 |
| Consensus MFE | -35.50 |
| Energy contribution | -34.82 |
| Covariance contribution | -0.68 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.761063 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13060806 101 - 23771897 GUCCGACAAUUGGGUUAAAGGACCCAUGGGCGUCGGCCUGUUGGCAUAUAUCUGUUCCGGAGACUGUUGCGCCAUUAAGCACGAGCACAGCGGCCUAUCCU (((((.....((((((....)))))))))))...((((.((((((.............(....)..(((.((......)).))))).))))))))...... ( -32.00) >DroSec_CAF1 48480 101 - 1 GUCCGACAAUUGGGUUAAAGGACCCAUGGGCGUAGGCCUGCUGGCAUAUAUCUGCUCCGGCGACUGUUGUGCCAUUAAGCACGAGCACAGCGGCCUAUCCU (((((.....((((((....)))))))))))(((((((.((((((.......(((....)))....((((((......)))))))).)))))))))))... ( -41.60) >DroSim_CAF1 44274 101 - 1 GUCCGACAAUUGGGUUAAAGGACCCAUGGGCGUCGGCCUGCUGGCAUAUAUCUGCUCCGGCGACUGUUGUGCUAUUAAGCACGAGCACAGCGGCCUAUCCU (((((.....((((((....)))))))))))...((((.((((((.......(((....)))....(((((((....))))))))).))))))))...... ( -39.80) >DroEre_CAF1 45172 101 - 1 GUCCGAGAAUUGGGUCAAGGGACCCAUGGGCGUUGGUCUGCUGGCCUAUAUCUGCUCCGGGGACUGUUGUGCCAUGAAGCACGAGCACAGCGGCUUAUCCU ..(((.....((((((....)))))).(((((..((((....))))......))))))))(((((((((((((.((...)).).)))))))))....))). ( -35.90) >DroYak_CAF1 47047 101 - 1 GUCCGAGAAUUGGGUCAAGGGACCCAUGGGCGUUGGUCUGCUGGCAUAUAUCUGCUCCGGAGACUGUUGUGCCAUCAAGCACGAGCACAGCGGCUUAUCCU (((((.....((((((....)))))))))))((..(....)..)).............(((..(((((((((..((......)))))))))))....))). ( -35.30) >consensus GUCCGACAAUUGGGUUAAAGGACCCAUGGGCGUCGGCCUGCUGGCAUAUAUCUGCUCCGGAGACUGUUGUGCCAUUAAGCACGAGCACAGCGGCCUAUCCU (((((.....((((((....)))))))))))...((((.((((((.............(....)..((((((......)))))))).))))))))...... (-35.50 = -34.82 + -0.68)



| Location | 13,060,833 – 13,060,941 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 90.56 |
| Mean single sequence MFE | -36.94 |
| Consensus MFE | -28.26 |
| Energy contribution | -29.50 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.637194 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13060833 108 + 23771897 UGGCGCAACAGUCUCCGGAACAGAUAUAUGCCAACAGGCCGACGCCCAUGGGUCCUUUAACCCAAUUGUCGGACGGGAAACCCGGGCGAACCUUUCCGGGACUUCCAU (((......((((.((((((.((......(((....)))...(((((.(((((......)))))..........((....)).)))))...)))))))))))).))). ( -39.90) >DroSec_CAF1 48507 108 + 1 UGGCACAACAGUCGCCGGAGCAGAUAUAUGCCAGCAGGCCUACGCCCAUGGGUCCUUUAACCCAAUUGUCGGACAGGAAGCCCGGGCGAACCUUUCCGGGACUUCCAU .(((((....)).)))............((((.((((((....)))..(((((......)))))..))).)).))(((((((((((........)))))).))))).. ( -37.70) >DroSim_CAF1 44301 108 + 1 UAGCACAACAGUCGCCGGAGCAGAUAUAUGCCAGCAGGCCGACGCCCAUGGGUCCUUUAACCCAAUUGUCGGACAGGAAGCCCGGGCGAACCUUUCCGGGACUUCCAU ..........((((((.(.(((......))))....)))(((((....(((((......)))))..)))))))).(((((((((((........)))))).))))).. ( -40.90) >DroEre_CAF1 45199 108 + 1 UGGCACAACAGUCCCCGGAGCAGAUAUAGGCCAGCAGACCAACGCCCAUGGGUCCCUUGACCCAAUUCUCGGACAGGAGGCCCAAGCGAUCCUUUCCAGGACUUCCAU (((......(((((..((((..(((...((........))..(((...((((((.((((.((........)).)))).)))))).))))))..)))).))))).))). ( -32.30) >DroYak_CAF1 47074 108 + 1 UGGCACAACAGUCUCCGGAGCAGAUAUAUGCCAGCAGACCAACGCCCAUGGGUCCCUUGACCCAAUUCUCGGACAGGAGAACCAAGCGAUCCUUUCCGGGACUUCCAU .(((......((((.(.(.(((......)))).).))))....)))..((((((....))))))..(((((((.((((...........)))).)))))))....... ( -33.90) >consensus UGGCACAACAGUCGCCGGAGCAGAUAUAUGCCAGCAGGCCAACGCCCAUGGGUCCUUUAACCCAAUUGUCGGACAGGAAGCCCGGGCGAACCUUUCCGGGACUUCCAU .........((((.((((((.((......(((....)))...(((((.(((((......)))))...........((....)))))))...))))))))))))..... (-28.26 = -29.50 + 1.24)

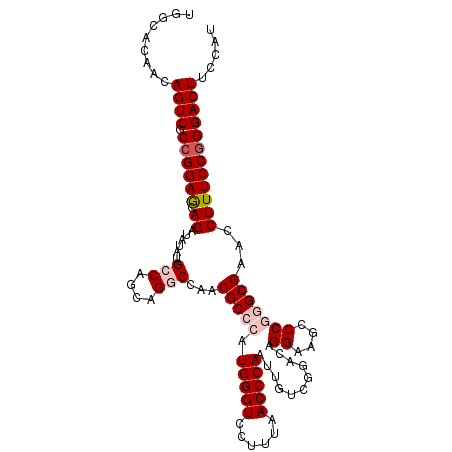

| Location | 13,060,833 – 13,060,941 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 90.56 |
| Mean single sequence MFE | -43.72 |
| Consensus MFE | -41.12 |
| Energy contribution | -39.92 |
| Covariance contribution | -1.20 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.09 |
| SVM RNA-class probability | 0.987824 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13060833 108 - 23771897 AUGGAAGUCCCGGAAAGGUUCGCCCGGGUUUCCCGUCCGACAAUUGGGUUAAAGGACCCAUGGGCGUCGGCCUGUUGGCAUAUAUCUGUUCCGGAGACUGUUGCGCCA .(((.((((((((((((((..(((((((((...((((((.....((((((....))))))))))))..))))))..)))....)))).)))))).))))......))) ( -43.60) >DroSec_CAF1 48507 108 - 1 AUGGAAGUCCCGGAAAGGUUCGCCCGGGCUUCCUGUCCGACAAUUGGGUUAAAGGACCCAUGGGCGUAGGCCUGCUGGCAUAUAUCUGCUCCGGCGACUGUUGUGCCA .(((.(((((((((.((((.(((((((((.....))))......((((((....)))))).)))))...(((....)))....))))..))))).))))......))) ( -45.90) >DroSim_CAF1 44301 108 - 1 AUGGAAGUCCCGGAAAGGUUCGCCCGGGCUUCCUGUCCGACAAUUGGGUUAAAGGACCCAUGGGCGUCGGCCUGCUGGCAUAUAUCUGCUCCGGCGACUGUUGUGCUA (..(.(((((((((.((((..((((((((.....))))......((((((....)))))).))))(((((....)))))....))))..))))).)))).)..).... ( -45.30) >DroEre_CAF1 45199 108 - 1 AUGGAAGUCCUGGAAAGGAUCGCUUGGGCCUCCUGUCCGAGAAUUGGGUCAAGGGACCCAUGGGCGUUGGUCUGCUGGCCUAUAUCUGCUCCGGGGACUGUUGUGCCA .(((.(((((((((..((((....((((((..(.(.(((((...((((((....))))))....).)))).).)..)))))).))))..))).))))))......))) ( -42.30) >DroYak_CAF1 47074 108 - 1 AUGGAAGUCCCGGAAAGGAUCGCUUGGUUCUCCUGUCCGAGAAUUGGGUCAAGGGACCCAUGGGCGUUGGUCUGCUGGCAUAUAUCUGCUCCGGAGACUGUUGUGCCA .(((.(((((((((..((((.(((((((((((......)))))))(((((....)))))..))))((..(....)..))....))))..))))).))))......))) ( -41.50) >consensus AUGGAAGUCCCGGAAAGGUUCGCCCGGGCUUCCUGUCCGACAAUUGGGUUAAAGGACCCAUGGGCGUCGGCCUGCUGGCAUAUAUCUGCUCCGGAGACUGUUGUGCCA .(((.(((((((((.((((..(((((((((...((((((.....((((((....))))))))))))..))))))..)))....))))..))))).))))......))) (-41.12 = -39.92 + -1.20)



| Location | 13,060,873 – 13,060,975 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 89.71 |
| Mean single sequence MFE | -44.06 |
| Consensus MFE | -35.86 |
| Energy contribution | -37.66 |
| Covariance contribution | 1.80 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.22 |
| Structure conservation index | 0.81 |
| SVM decision value | 3.19 |
| SVM RNA-class probability | 0.998691 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13060873 102 + 23771897 GACGCCCAUGGGUCCUUUAACCCAAUUGUCGGACGGGAAACCCGGGCGAACCUUUCCGGGACUUCCAUGCCCAGGGAGCCUACUUAGUCCUUGGGCGGAAUG ..(((((.(((((......)))))..........((....)).))))).....((((.((....))..(((((((((..........))))))))))))).. ( -43.60) >DroSec_CAF1 48547 102 + 1 UACGCCCAUGGGUCCUUUAACCCAAUUGUCGGACAGGAAGCCCGGGCGAACCUUUCCGGGACUUCCAUGUCCAGGGAGCCUCCUUAGUCCUUGGGCGGAAUG ..(((((((((((......)))))......((((((((((((((((........)))))).))))).)))))((((.....))))......))))))..... ( -47.90) >DroSim_CAF1 44341 102 + 1 GACGCCCAUGGGUCCUUUAACCCAAUUGUCGGACAGGAAGCCCGGGCGAACCUUUCCGGGACUUCCAUGUCCAGGGAGCCUCCUUAGUCCUUGGGCAGAAUG ...((((((((((......)))))......((((((((((((((((........)))))).))))).)))))((((.....))))......)))))...... ( -45.70) >DroEre_CAF1 45239 102 + 1 AACGCCCAUGGGUCCCUUGACCCAAUUCUCGGACAGGAGGCCCAAGCGAUCCUUUCCAGGACUUCCAUGUCCAGGGAGCCUCUUUGUUCCUUGGGCGGAGUG ..((((((((((((....))))))......(((((((((((((..(.((.....))).((((......))))..)).))))).))))))..))))))..... ( -45.00) >DroYak_CAF1 47114 102 + 1 AACGCCCAUGGGUCCCUUGACCCAAUUCUCGGACAGGAGAACCAAGCGAUCCUUUCCGGGACUUCCAUGUCCAGGUAGCCUCUUUACUCCUUGGGCGGAAUG ..((((((((((((....)))))).....((((.((((...........)))).))))((((......))))(((.((........)))))))))))..... ( -38.10) >consensus AACGCCCAUGGGUCCUUUAACCCAAUUGUCGGACAGGAAGCCCGGGCGAACCUUUCCGGGACUUCCAUGUCCAGGGAGCCUCCUUAGUCCUUGGGCGGAAUG ..(((((.(((((......)))))......((((((((((((((((........)))))).))))).)))))(((((..........))))))))))..... (-35.86 = -37.66 + 1.80)


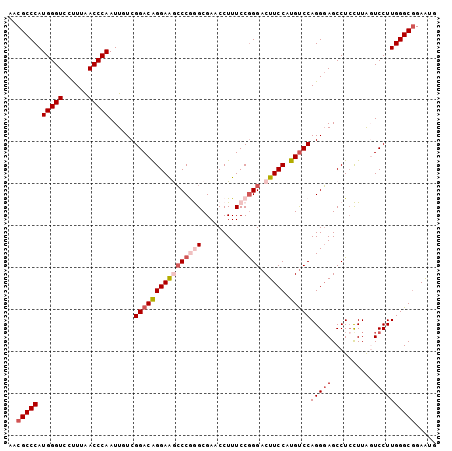
| Location | 13,060,873 – 13,060,975 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 89.71 |
| Mean single sequence MFE | -44.38 |
| Consensus MFE | -40.98 |
| Energy contribution | -41.30 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.92 |
| SVM decision value | 4.59 |
| SVM RNA-class probability | 0.999925 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13060873 102 - 23771897 CAUUCCGCCCAAGGACUAAGUAGGCUCCCUGGGCAUGGAAGUCCCGGAAAGGUUCGCCCGGGUUUCCCGUCCGACAAUUGGGUUAAAGGACCCAUGGGCGUC .....((((((.((((.......(((.....)))..(((((.(((((..........)))))))))).))))......((((((....)))))))))))).. ( -42.90) >DroSec_CAF1 48547 102 - 1 CAUUCCGCCCAAGGACUAAGGAGGCUCCCUGGACAUGGAAGUCCCGGAAAGGUUCGCCCGGGCUUCCUGUCCGACAAUUGGGUUAAAGGACCCAUGGGCGUA .....((((((.((((..(((((((((((.((((......)))).))...((....)).)))))))))))))......((((((....)))))))))))).. ( -48.60) >DroSim_CAF1 44341 102 - 1 CAUUCUGCCCAAGGACUAAGGAGGCUCCCUGGACAUGGAAGUCCCGGAAAGGUUCGCCCGGGCUUCCUGUCCGACAAUUGGGUUAAAGGACCCAUGGGCGUC ......(((((.((((..(((((((((((.((((......)))).))...((....)).)))))))))))))......((((((....)))))))))))... ( -47.00) >DroEre_CAF1 45239 102 - 1 CACUCCGCCCAAGGAACAAAGAGGCUCCCUGGACAUGGAAGUCCUGGAAAGGAUCGCUUGGGCCUCCUGUCCGAGAAUUGGGUCAAGGGACCCAUGGGCGUU .....((((((.(((.((..(((((((((.((((......)))).)).(((.....)))))))))).)))))......((((((....)))))))))))).. ( -44.40) >DroYak_CAF1 47114 102 - 1 CAUUCCGCCCAAGGAGUAAAGAGGCUACCUGGACAUGGAAGUCCCGGAAAGGAUCGCUUGGUUCUCCUGUCCGAGAAUUGGGUCAAGGGACCCAUGGGCGUU .....((((((((((((......))).)))((((......))))((((.((((..((...))..)))).)))).....((((((....)))))))))))).. ( -39.00) >consensus CAUUCCGCCCAAGGACUAAGGAGGCUCCCUGGACAUGGAAGUCCCGGAAAGGUUCGCCCGGGCUUCCUGUCCGACAAUUGGGUUAAAGGACCCAUGGGCGUC .....((((((.((((..(((((((((((.((((......)))).))...((....)).)))))))))))))......((((((....)))))))))))).. (-40.98 = -41.30 + 0.32)

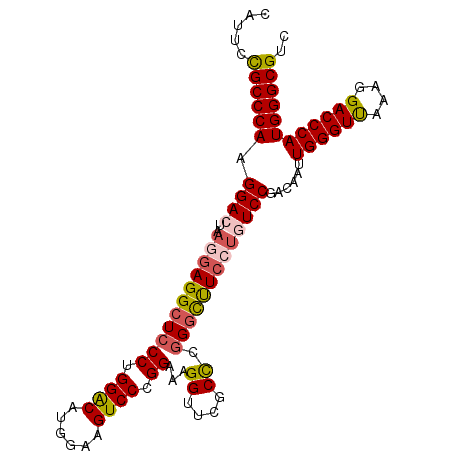
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:26:04 2006