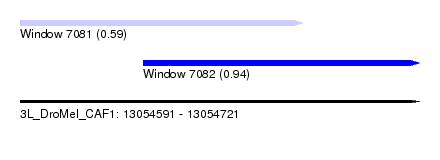
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,054,591 – 13,054,721 |
| Length | 130 |
| Max. P | 0.935514 |

| Location | 13,054,591 – 13,054,683 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.47 |
| Mean single sequence MFE | -27.05 |
| Consensus MFE | -20.71 |
| Energy contribution | -21.43 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.587540 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
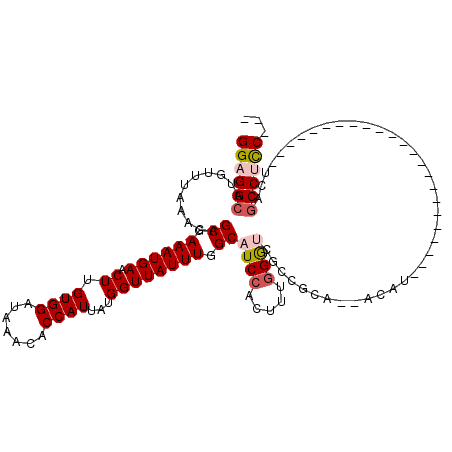
>3L_DroMel_CAF1 13054591 92 + 23771897 GGAGGCAUGUUUAAACAGCGAAAUGAACUUGUGGAUAAACACCAUUAUGGUUAUUUGGCAUCCACUUUGGGUCGCCGCA--ACAU-----------------------UCAGCCUUC--- ((((((.(((....)))((.((((((.((.((((.......))))...))))))))(((((((.....)))).))))).--....-----------------------...))))))--- ( -25.90) >DroPse_CAF1 33806 85 + 1 GGAGGCAUGUUUAAACAGCGAAAUGAACUUGUGGAUAAACACCAUUAUGGUUAUUUGGCAUCCACUUUAGG---CGGCA--AC---------------------------AGCCACC--- ((.(((..(((((((..((.((((((.((.((((.......))))...)))))))).))......))))))---)(...--.)---------------------------.))).))--- ( -21.00) >DroSim_CAF1 38141 92 + 1 GGAGGCAUGUUUAAACAGCGAAAUGAACUUGUGGAUAAACACCAUUAUGGUUAUUUGGCAUCCACUUUGGGUCGCCGCA--ACAU-----------------------UCAGCCUCC--- ((((((.(((....)))((.((((((.((.((((.......))))...))))))))(((((((.....)))).))))).--....-----------------------...))))))--- ( -28.80) >DroEre_CAF1 38927 92 + 1 GGAGGCAUGUUUAAACAGCGAAAUGAACUUGUGGAUAAACACCAUUAUGGUUAUUUGGCAUCCACUUUGGGUCGCCGCA--GCAU-----------------------UCAGCCUCC--- ((((((.(((....)))((.((((((.((.((((.......))))...))))))))(((((((.....)))).))))).--....-----------------------...))))))--- ( -28.80) >DroYak_CAF1 40634 92 + 1 GGAGGCAUGUUUAAACAGCGAAAUGAACUUGUGGAUAAACACCAUUAUGGUUAUUUGGCAUCCACUUUGGGUCGCCGCA--ACAU-----------------------UCAGCCUCC--- ((((((.(((....)))((.((((((.((.((((.......))))...))))))))(((((((.....)))).))))).--....-----------------------...))))))--- ( -28.80) >DroAna_CAF1 37509 120 + 1 GGAGGCAUGUUUAAACAGCGAAAUGAACUUGUGGAUAAACACCAUUAUGGUUAUUUGGCAUCCACUUUGGACAGCCACCGACCAUCGCCACCGCCAACCGCAACCAUCUCAACCGCCACU ...(((.(((....)))..((.(((...((((((............((((((...((((.(((.....)))..))))..)))))).((....))...)))))).))).))....)))... ( -29.00) >consensus GGAGGCAUGUUUAAACAGCGAAAUGAACUUGUGGAUAAACACCAUUAUGGUUAUUUGGCAUCCACUUUGGGUCGCCGCA__ACAU_______________________UCAGCCUCC___ ((((((...........((.((((((.((.((((.......))))...)))))))).))((((.....)))).......................................))))))... (-20.71 = -21.43 + 0.72)


| Location | 13,054,631 – 13,054,721 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 93.69 |
| Mean single sequence MFE | -23.98 |
| Consensus MFE | -21.98 |
| Energy contribution | -22.42 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.935514 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13054631 90 + 23771897 ACCAUUAUGGUUAUUUGGCAUCCACUUUGGGUCGCCGCAACAUUCAGCCUUCGAUGUUUGUCGCCCUACAGGGUAUGCCGACUACCCCA-C ........(((...(((((((((.((.((((.((.((.((((((........)))))))).)).)))).)))).)))))))..)))...-. ( -26.70) >DroSec_CAF1 42324 90 + 1 ACCAUUAUGGUUAUUUGGCAUCCACUUUGGGUCGCCGCAACAUUCAGCCUCCGAUGUUUGUCGCCCUACAGGGUAUGCCGACCACCCCA-C ........(((...(((((((((.((.((((.((.((.((((((........)))))))).)).)))).)))).)))))))..)))...-. ( -26.00) >DroSim_CAF1 38181 90 + 1 ACCAUUAUGGUUAUUUGGCAUCCACUUUGGGUCGCCGCAACAUUCAGCCUCCGAUGUUUGUCGCCCUACAGGGUAUGCCGACCACCCCA-C ........(((...(((((((((.((.((((.((.((.((((((........)))))))).)).)))).)))).)))))))..)))...-. ( -26.00) >DroEre_CAF1 38967 83 + 1 ACCAUUAUGGUUAUUUGGCAUCCACUUUGGGUCGCCGCAGCAUUCAGCCUCCGAUGUUUGUCGCCCUACAGGGUAUACCGACC-------- .......((((.....(((((((.....)))).)))((((((((........)))).)))).((((....))))..))))...-------- ( -19.90) >DroYak_CAF1 40674 91 + 1 ACCAUUAUGGUUAUUUGGCAUCCACUUUGGGUCGCCGCAACAUUCAGCCUCCUAUGUUUGUCGCCCUACAGGGUAUACCGACCACUCCAGC .......(((((....(((((((.....)))).)))...(((..((........))..))).((((....)))).....)))))....... ( -21.30) >consensus ACCAUUAUGGUUAUUUGGCAUCCACUUUGGGUCGCCGCAACAUUCAGCCUCCGAUGUUUGUCGCCCUACAGGGUAUGCCGACCACCCCA_C (((.....)))...(((((((((.((.((((.((.((.((((((........)))))))).)).)))).)))).))))))).......... (-21.98 = -22.42 + 0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:25:55 2006