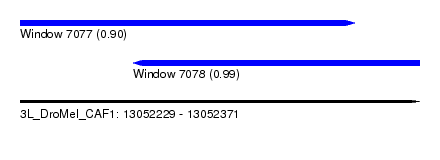
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,052,229 – 13,052,371 |
| Length | 142 |
| Max. P | 0.985793 |

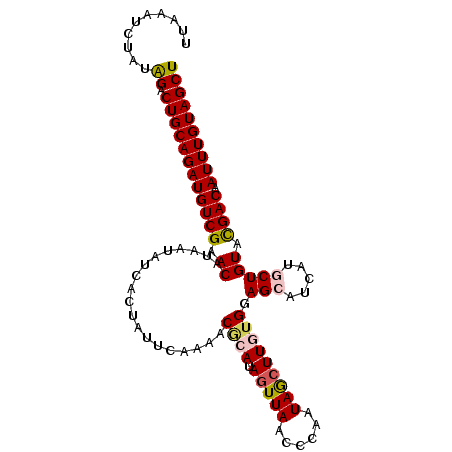
| Location | 13,052,229 – 13,052,348 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 93.78 |
| Mean single sequence MFE | -34.70 |
| Consensus MFE | -29.82 |
| Energy contribution | -29.98 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.900714 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13052229 119 + 23771897 GAUUGGCGUGGCGUAGGUUGUGGCAACAAUUUACGCUACGAGCUACAAAUUGUCAUACAGCAUGAUGCUCCACAAGCUAUUGGGUUAACUAUGCGUUUUGAAUAGUGAUAUUAUGUUCG .......((((((((((((((....))))))))))))))((((.......((((((....((.(((((......((((....))))......))))).))....))))))....)))). ( -36.00) >DroSec_CAF1 39971 119 + 1 GAUUGGCGUGGCGUAGGUUGUGGCAACAAUUUACGCUACGAGCUACAAAUUGUCGUACAGCAUGAUGCUCCACAAGCUAUUGGGUUAACUAUGCGUUUUGAAUAGUGAUAUUAUGUUCG .......((((((((((((((....))))))))))))))((((.....(((((((....(((((.....((.(((....))))).....)))))....)).)))))........)))). ( -34.02) >DroSim_CAF1 35820 119 + 1 GAUUGGCGUGGCGUAGGUGGUGGCAACAAUUUACGCUACGAGCUACAAAUUGUCGUACAGCAUGAUGCUCCACAAGCUAUUGUGUUAACUAUGCGUUUUGAAUAGUGAUAUUAUGUUCG .......(((((((((((..((....)))))))))))))((((..((((.((.((((.((((..(((((.....))).))..))))...)))))).))))((((....))))..)))). ( -33.70) >DroEre_CAF1 36426 117 + 1 GAUUGGCGUGGCGUAGGUUGUGGCCACAAUUUACGCUACGAGCUACAAAUUGUCGUACAGCACGAUUCUCC--AAGUUAUUGGGUUAAGUAUGCGUUUUGCAUAGUGAUAUUAUGUUCG .......((((((((((((((....))))))))))))))((((........(((((.....)))))...((--((....))))((((..(((((.....))))).)))).....)))). ( -35.10) >DroYak_CAF1 38036 119 + 1 GAUUGGCGUGGCGUAGGUUGUGGCUGCAAUUUGCGCUACGAGCUACAAAUUGUCGUACAGCAUGAUUCUCCAUAAGUUAGUGGGUUAACUAUGUGUUUUGAAUAGUGAUAUUGUGUUCG .......((((((((((((((....))))))))))))))((((.((((.(((.....)))(((.((((..((((((((((....)))))).))))....)))).)))...)))))))). ( -34.70) >consensus GAUUGGCGUGGCGUAGGUUGUGGCAACAAUUUACGCUACGAGCUACAAAUUGUCGUACAGCAUGAUGCUCCACAAGCUAUUGGGUUAACUAUGCGUUUUGAAUAGUGAUAUUAUGUUCG .......((((((((((((((....))))))))))))))((((.....(((((((....(((((....((((........)))).....)))))....)).)))))........)))). (-29.82 = -29.98 + 0.16)

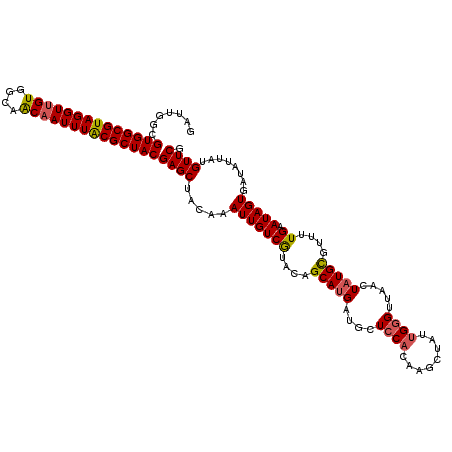
| Location | 13,052,269 – 13,052,371 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 93.53 |
| Mean single sequence MFE | -22.82 |
| Consensus MFE | -18.52 |
| Energy contribution | -19.04 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.81 |
| SVM decision value | 2.02 |
| SVM RNA-class probability | 0.985793 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13052269 102 - 23771897 UUAAAUCUAUAGACUGCAGAUGUCGAACAUAAUAUCACUAUUCAAAACGCAUAGUUAACCCAAUAGCUUGUGGAGCAUCAUGCUGUAUGACAAUUUGUAGCU ..........((.((((((((((((.((...................((((.(((((......))))))))).(((.....))))).)))).)))))))))) ( -22.60) >DroSec_CAF1 40011 102 - 1 UUAAAUCUAUAGACUGCAGAUGUCGAACAUAAUAUCACUAUUCAAAACGCAUAGUUAACCCAAUAGCUUGUGGAGCAUCAUGCUGUACGACAAUUUGUAGCU ..........((.((((((((((((.((...................((((.(((((......))))))))).(((.....))))).)))).)))))))))) ( -24.20) >DroSim_CAF1 35860 102 - 1 UUAAAUCUAUAGACUGCAGAUGUCGAACAUAAUAUCACUAUUCAAAACGCAUAGUUAACACAAUAGCUUGUGGAGCAUCAUGCUGUACGACAAUUUGUAGCU ..........((.((((((((((((.((...................((((.(((((......))))))))).(((.....))))).)))).)))))))))) ( -24.20) >DroEre_CAF1 36466 100 - 1 UUAAAUCUACGGACUGCAGAUGUCGAACAUAAUAUCACUAUGCAAAACGCAUACUUAACCCAAUAACUU--GGAGAAUCGUGCUGUACGACAAUUUGUAGCU .....((....))((((((((((((.(((......((((((((.....)))))......((((....))--))......))).))).)))).)))))))).. ( -23.60) >DroYak_CAF1 38076 102 - 1 UUAAAUCUAAAGACUGCAGAUGUCGAACACAAUAUCACUAUUCAAAACACAUAGUUAACCCACUAACUUAUGGAGAAUCAUGCUGUACGACAAUUUGUAGCU ..........((.((((((((((((.(((((..................((((((((......)))).))))(.....).)).))).)))).)))))))))) ( -19.50) >consensus UUAAAUCUAUAGACUGCAGAUGUCGAACAUAAUAUCACUAUUCAAAACGCAUAGUUAACCCAAUAGCUUGUGGAGCAUCAUGCUGUACGACAAUUUGUAGCU ..........((.((((((((((((.((...................((((.(((((......))))))))).(((.....))))).)))).)))))))))) (-18.52 = -19.04 + 0.52)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:25:51 2006