| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,042,210 – 13,042,305 |
| Length | 95 |
| Max. P | 0.784525 |

| Location | 13,042,210 – 13,042,305 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
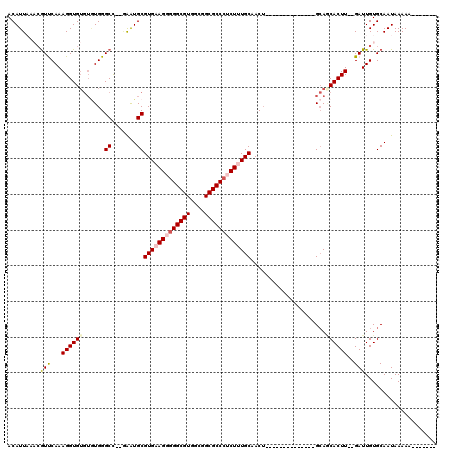
| Mean pairwise identity | 82.13 |
| Mean single sequence MFE | -34.53 |
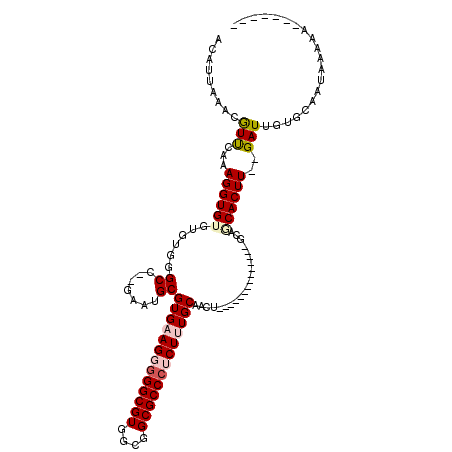
| Consensus MFE | -23.34 |
| Energy contribution | -24.07 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.784525 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13042210 95 - 23771897 ACAUUAAACGUUCAAAGGUGUGUAUGGGCC--GAAUGCGUGAAGGGGGCGUGGCGGCGCCCUCUUUGCAACU--------------GCAGCACUU--GAUUGUGCAAUAAAAA------- ........(((((...(((.(....).)))--))))).(..((((((((((....))))))))))..)....--------------...((((..--....))))........------- ( -33.40) >DroPse_CAF1 22699 107 - 1 ACAUUUUCCGUUCAAAGGUG----UGGGCC--CGAUGCGUGAAGUGGGCGUGCCGGCGCCCUCUCUGCCACACGGUGGAACACAGGGGAACACUUGUGAUUGUGCAAUAAAAA------- .........((((....(((----((((((--((..((((.......))))..))).))))..((..((....))..))))))....))))..((((......))))......------- ( -35.10) >DroSec_CAF1 25712 95 - 1 ACAUUAAACGUUCAAAGGUGUGUGUGGGCC--AAAUGCGUGAAGGGGGCGUGGCGGCGCCCUCUUUGCAACU--------------GCAGCACUU--GAUUGUGCAAUAAAAA------- ..(((..(((.(((..(((((.((..(((.--....))(..((((((((((....))))))))))..)..).--------------.))))))))--)).)))..))).....------- ( -35.20) >DroEre_CAF1 26267 94 - 1 ACAUUAAACAUCCAAAGGUGUGUGUGGGCU--GAAUGCGUGAAGG-GGCGUGGCGGCGCCCUCUUUGCAACU--------------GCACCACUU--GAUUGUGCAAUAAAAA------- ..(((..(((.....(((((.(((..(((.--....))(..((((-(((((....)))))).)))..)..).--------------.))))))))--...)))..))).....------- ( -34.10) >DroYak_CAF1 27618 95 - 1 ACAUUAAACGUCCAAAGGUGUGUGUGGGCU--GAAUGCGUGAAGGGGGCGUGGCGGCGCCCUCUCUGCAACU--------------GCAGCACUU--GAUUGUGCAAUAAAAA------- .........(..(((((((((.((..(((.--....))((..(((((((((....)))))))))..))..).--------------.))))))))--..)))..)........------- ( -33.60) >DroAna_CAF1 25740 104 - 1 ACAUUUUCAGUUCAAAGGUGUGUGUGGGCUUGAAGUGCGUGAAGUGGGCGUGUCGGCGCCCUCUUUGCAACU--------------GCAACACUU--GAUUGUGCAAUAAAAAAUGUAAA (((((((..(((((.((((((.((..(((.......))(..(((.((((((....)))))).)))..)..).--------------.))))))))--.....)).)))..)))))))... ( -35.80) >consensus ACAUUAAACGUUCAAAGGUGUGUGUGGGCC__GAAUGCGUGAAGGGGGCGUGGCGGCGCCCUCUUUGCAACU______________GCAGCACUU__GAUUGUGCAAUAAAAA_______ .........(((...((((((......((.......))(((((((((((((....))))))))))))).....................))))))..))).................... (-23.34 = -24.07 + 0.72)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:25:46 2006