
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,038,478 – 13,038,569 |
| Length | 91 |
| Max. P | 0.729437 |

| Location | 13,038,478 – 13,038,569 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 77.49 |
| Mean single sequence MFE | -21.40 |
| Consensus MFE | -11.08 |
| Energy contribution | -12.75 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.683545 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
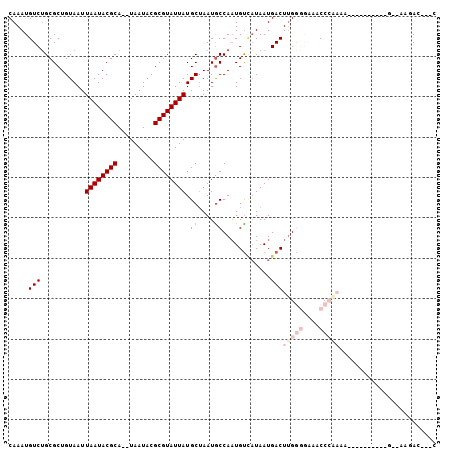
>3L_DroMel_CAF1 13038478 91 + 23771897 CAAAUGUCUGCGCUGUAAUUAAUACGCA--UAAUACGCGUAUUAUGCUAAUGCCAAUGUCAUAAUGACUCGAGGAAACCCGAAA---------UG-AAAGAC---G .....(((((((..(((..((((((((.--......)))))))))))...)))....(((.....)))(((.(....).)))..---------..-..))))---. ( -21.80) >DroVir_CAF1 25365 95 + 1 CAAAUGUCGGCGCUGUAAUUAAUACGCA--UAAUACGCGUAUUAUGCUAAUGCCAAUGCUUUAAUGACUUGG-----CCCAAAACUUCGUCGUGG-AAAGAC---A ....((((((((..(((..((((((((.--......)))))))))))...)))).....(((.(((((..((-----.......))..))))).)-)).)))---) ( -23.90) >DroPse_CAF1 19622 83 + 1 CAAAUGUCUGCGCUGUAAUUAAUACGCAUAUAAUACGCGUAUUAUGCUAAUGCCAAUGUCAUAAUGACUUGGGGAAACCG--------------------AA---C .........(((..(((..((((((((.........)))))))))))...)))(((.(((.....))))))((....)).--------------------..---. ( -19.20) >DroGri_CAF1 43491 96 + 1 CAAAUGUCUGCGCUGUAAUUAAUACGCA--UAAUACGCGUAUUAUGCAAAUGCCAUUGUAAUAAUGACUUGG-----CCCAAAACUUGCCCUUGGGUAAGAC---A .............((((..((((((((.--......))))))))))))...((((..((.......)).)))-----)......((((((....))))))..---. ( -23.60) >DroAna_CAF1 21909 96 + 1 CAAAUGUCUGCGCUGUAAUUAAUACGCA--UAAUACGCGUAUUAUGCUAAUACCAAUGUCAUAAUGACUUGAGGAAACCCAAAAAC-------UG-GAAGAGACUC .....((((.....(((..((((((((.--......))))))))))).....(((..(((.....)))(((.(....).)))....-------))-)...)))).. ( -20.70) >DroPer_CAF1 19579 83 + 1 CAAAUGUCUGCGCUGUAAUUAAUACGCAUAUAAUACGCGUAUUAUGCUAAUGCCAAUGUCAUAAUGACUUGGGGAAACCG--------------------AA---C .........(((..(((..((((((((.........)))))))))))...)))(((.(((.....))))))((....)).--------------------..---. ( -19.20) >consensus CAAAUGUCUGCGCUGUAAUUAAUACGCA__UAAUACGCGUAUUAUGCUAAUGCCAAUGUCAUAAUGACUUGGGGAAACCCAAAA__________G__AAGAC___C .....(((...........((((((((.........)))))))).((....))............)))(((((....)))))........................ (-11.08 = -12.75 + 1.67)


| Location | 13,038,478 – 13,038,569 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 77.49 |
| Mean single sequence MFE | -22.10 |
| Consensus MFE | -13.49 |
| Energy contribution | -13.35 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.729437 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
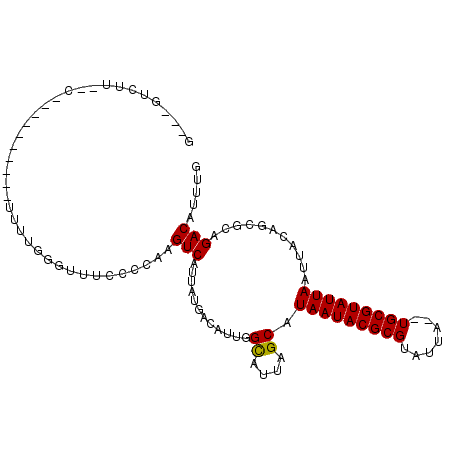
>3L_DroMel_CAF1 13038478 91 - 23771897 C---GUCUUU-CA---------UUUCGGGUUUCCUCGAGUCAUUAUGACAUUGGCAUUAGCAUAAUACGCGUAUUA--UGCGUAUUAAUUACAGCGCAGACAUUUG .---((((..-..---------..(((((....)))))(((.....)))....((....((((((((....)))))--)))(((.....)))...))))))..... ( -23.10) >DroVir_CAF1 25365 95 - 1 U---GUCUUU-CCACGACGAAGUUUUGGG-----CCAAGUCAUUAAAGCAUUGGCAUUAGCAUAAUACGCGUAUUA--UGCGUAUUAAUUACAGCGCCGACAUUUG .---(((...-....))).((((.(((((-----((((((.......)).))))).......((((((((((...)--))))))))).........)))).)))). ( -22.80) >DroPse_CAF1 19622 83 - 1 G---UU--------------------CGGUUUCCCCAAGUCAUUAUGACAUUGGCAUUAGCAUAAUACGCGUAUUAUAUGCGUAUUAAUUACAGCGCAGACAUUUG (---((--------------------(((((...(((((((.....))).))))....))).(((((((((((...))))))))))).......))..)))..... ( -19.80) >DroGri_CAF1 43491 96 - 1 U---GUCUUACCCAAGGGCAAGUUUUGGG-----CCAAGUCAUUAUUACAAUGGCAUUUGCAUAAUACGCGUAUUA--UGCGUAUUAAUUACAGCGCAGACAUUUG (---((((..(((((((.....)))))))-----....((((((.....)))))).......((((((((((...)--)))))))))..........))))).... ( -24.60) >DroAna_CAF1 21909 96 - 1 GAGUCUCUUC-CA-------GUUUUUGGGUUUCCUCAAGUCAUUAUGACAUUGGUAUUAGCAUAAUACGCGUAUUA--UGCGUAUUAAUUACAGCGCAGACAUUUG ..(((((..(-((-------((..(((((....)))))(((.....))))))))........((((((((((...)--)))))))))........).))))..... ( -22.50) >DroPer_CAF1 19579 83 - 1 G---UU--------------------CGGUUUCCCCAAGUCAUUAUGACAUUGGCAUUAGCAUAAUACGCGUAUUAUAUGCGUAUUAAUUACAGCGCAGACAUUUG (---((--------------------(((((...(((((((.....))).))))....))).(((((((((((...))))))))))).......))..)))..... ( -19.80) >consensus G___GUCUU__C__________UUUUGGGUUUCCCCAAGUCAUUAUGACAUUGGCAUUAGCAUAAUACGCGUAUUA__UGCGUAUUAAUUACAGCGCAGACAUUUG ......................................(((............((....)).(((((((((.......)))))))))...........)))..... (-13.49 = -13.35 + -0.14)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:25:43 2006