
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,016,868 – 13,016,958 |
| Length | 90 |
| Max. P | 0.994714 |

| Location | 13,016,868 – 13,016,958 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 73.01 |
| Mean single sequence MFE | -32.23 |
| Consensus MFE | -22.95 |
| Energy contribution | -23.98 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.71 |
| SVM decision value | 2.50 |
| SVM RNA-class probability | 0.994714 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
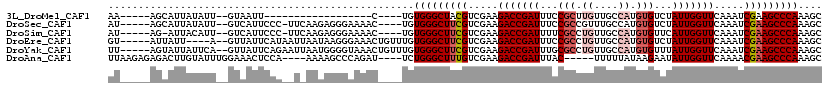
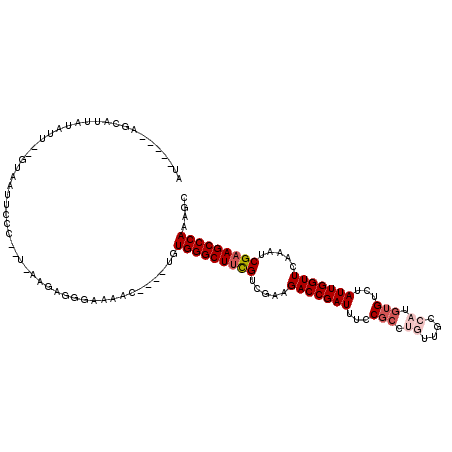
>3L_DroMel_CAF1 13016868 90 + 23771897 AA-----AGCAUUAUAUU--GUAAUU------------------C----UGUGGGCUACGUCGAAGACCGAUUUCCGCUUGUUGCCAUGUGUCUAUUGGUUCAAAUCGAAGCCCAAAGC ..-----.(((......)--))....------------------(----(.((((((.((.....(((((((...(((.((....)).)))...))))))).....)).)))))).)). ( -23.60) >DroSec_CAF1 728 107 + 1 AU-----AGCAUUAUAUU--GUCAUUCCC-UUCAAGAGGGAAAAC----UGUGGGCUUCGUCGAAGACCGAUUUCCGCCGUUUGCCAUGUGUCUAUUGGUUCAAAUCGAAGCCCAAAGC ..-----.(((......)--))..(((((-((...)))))))..(----(.(((((((((.....(((((((...(((..........)))...))))))).....))))))))).)). ( -33.40) >DroSim_CAF1 729 106 + 1 AU-----AG-AUUACAUU--GUCAUUCCC-UUCAAGAGGGAAAAC----UGUGGGCUUCGUCGAAGACCGAUUUUCGCCUGUUGCCAUGUGUUCAUUGGUUCAAAUCGAAGCCCAAAGC ..-----.(-((......--))).(((((-((...)))))))..(----(.(((((((((.....(((((((...(((.((....)).)))...))))))).....))))))))).)). ( -36.40) >DroEre_CAF1 918 108 + 1 GU-----AUUAUU----A--GUUAUUCAUAAUUAAUAAGGGAAACUGUUUGUGGGCUUCGUCGAAGACCGAUUUCCGCCUGUUGCCAUGUGUCUAUUGGUUCAAAUCGAAGCCCAAAGC ..-----.(((((----(--((((....))))))))))((....))((((.(((((((((.....(((((((...(((.((....)).)))...))))))).....))))))))))))) ( -34.80) >DroYak_CAF1 943 112 + 1 UU-----AGUAUUAUUCA--GUUAUUCAGAAUUAAUGGGGUAAACUGUUUGUGGGCUUCGUCGAAGACCGAUUUGCGCCUGUUGCCAUGUGUUUAUUGGUUCAAAUCGAAGCCCAAAGC .(-----(((.((((((.--((((........)))).))))))))))....(((((((((.....(((((((..((((.((....)).))))..))))))).....))))))))).... ( -36.50) >DroAna_CAF1 601 106 + 1 UUAAGAGAGACUUGUAUUUGGAAACUCCA----AAAAGCCCAGAU----UCUGGGCUUUGUCGAAGACCGAUUUAC-----UUUUUAUAAGAAUAUUGGUUCAAAACGAAGCCCAAAGC .....((((.((.((.(((((.....)))----))..))..)).)----)))(((((((((....(((((((...(-----((.....)))...)))))))....)))))))))..... ( -28.70) >consensus AU_____AGCAUUAUAUU__GUAAUUCCC__U_AAGAGGGAAAAC____UGUGGGCUUCGUCGAAGACCGAUUUCCGCCUGUUGCCAUGUGUCUAUUGGUUCAAAUCGAAGCCCAAAGC ...................................................(((((((((.....(((((((...(((.((....)).)))...))))))).....))))))))).... (-22.95 = -23.98 + 1.03)
| Location | 13,016,868 – 13,016,958 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 73.01 |
| Mean single sequence MFE | -26.99 |
| Consensus MFE | -17.30 |
| Energy contribution | -18.47 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.972393 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
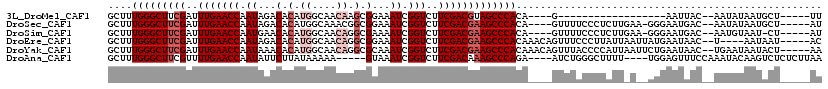
>3L_DroMel_CAF1 13016868 90 - 23771897 GCUUUGGGCUUCGAUUUGAACCAAUAGACACAUGGCAACAAGCGGAAAUCGGUCUUCGACGUAGCCCACA----G------------------AAUUAC--AAUAUAAUGCU-----UU .((.((((((((((...((.((.((...(.(.((....)).).)...)).)))).))))...)))))).)----)------------------......--...........-----.. ( -20.10) >DroSec_CAF1 728 107 - 1 GCUUUGGGCUUCGAUUUGAACCAAUAGACACAUGGCAAACGGCGGAAAUCGGUCUUCGACGAAGCCCACA----GUUUUCCCUCUUGAA-GGGAAUGAC--AAUAUAAUGCU-----AU ((..(((((((((..((((((((.........))).....(((.(....).)))))))))))))))))..----(((((((((.....)-))))).)))--........)).-----.. ( -31.90) >DroSim_CAF1 729 106 - 1 GCUUUGGGCUUCGAUUUGAACCAAUGAACACAUGGCAACAGGCGAAAAUCGGUCUUCGACGAAGCCCACA----GUUUUCCCUCUUGAA-GGGAAUGAC--AAUGUAAU-CU-----AU (((.(((((((((..(((((((.((.......((....)).......)).)))..))))))))))))).)----)).((((((.....)-)))))....--........-..-----.. ( -32.14) >DroEre_CAF1 918 108 - 1 GCUUUGGGCUUCGAUUUGAACCAAUAGACACAUGGCAACAGGCGGAAAUCGGUCUUCGACGAAGCCCACAAACAGUUUCCCUUAUUAAUUAUGAAUAAC--U----AAUAAU-----AC ....(((((((((..(((((............((....))(((.(....).))))))))))))))))).............((((((.(((....))).--)----))))).-----.. ( -24.50) >DroYak_CAF1 943 112 - 1 GCUUUGGGCUUCGAUUUGAACCAAUAAACACAUGGCAACAGGCGCAAAUCGGUCUUCGACGAAGCCCACAAACAGUUUACCCCAUUAAUUCUGAAUAAC--UGAAUAAUACU-----AA ....((((((((((((((..((..........((....))))..)))))).(((...)))))))))))....(((((...................)))--)).........-----.. ( -27.01) >DroAna_CAF1 601 106 - 1 GCUUUGGGCUUCGUUUUGAACCAAUAUUCUUAUAAAAA-----GUAAAUCGGUCUUCGACAAAGCCCAGA----AUCUGGGCUUUU----UGGAGUUUCCAAAUACAAGUCUCUCUUAA .....((((((.((((((.(((.((...(((.....))-----)...)).)))((((((.(((((((((.----..))))))))))----)))))....)))).))))))))....... ( -26.30) >consensus GCUUUGGGCUUCGAUUUGAACCAAUAGACACAUGGCAACAGGCGGAAAUCGGUCUUCGACGAAGCCCACA____GUUUUCCCUAUU_A__GGGAAUAAC__AAUAUAAUACU_____AU ....(((((((((..(((((((.((...(.(.((....)).).)...)).)))..)))))))))))))................................................... (-17.30 = -18.47 + 1.17)
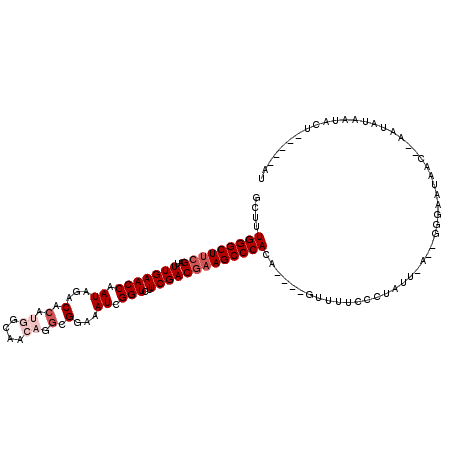
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:25:37 2006