
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,016,676 – 13,016,788 |
| Length | 112 |
| Max. P | 0.974810 |

| Location | 13,016,676 – 13,016,788 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 78.99 |
| Mean single sequence MFE | -31.30 |
| Consensus MFE | -24.14 |
| Energy contribution | -24.25 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.06 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.974810 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
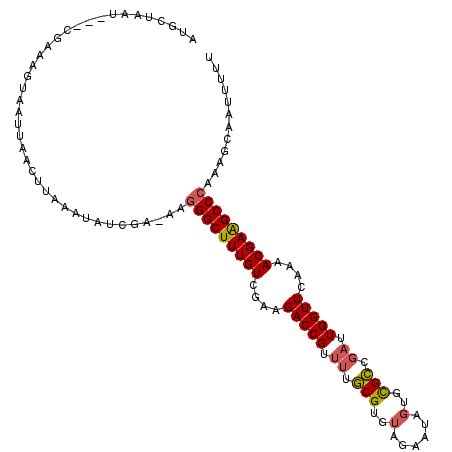
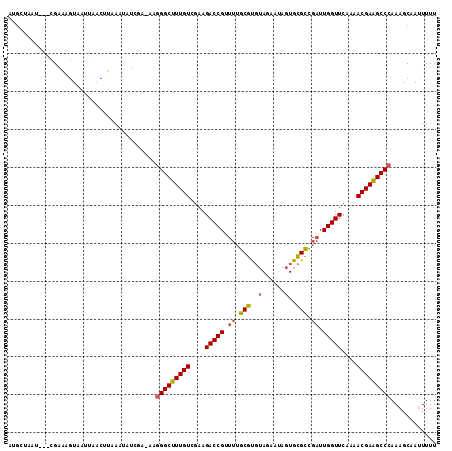
>3L_DroMel_CAF1 13016676 112 + 23771897 AUGCUAAUUACCGAAAGUAAUUAACUUAAAUAUCGA-AAGGGCUUUGUCGAAGACCGUUUUGCGUGUAGAAUAGUGCGCCGAUUGGUUCAAAACGAAGCCCAAAGCAAUUUUU .(((((((((.(....))))))).............-..(((((((((....(((((.((.(((..(......)..))).)).)))))....)))))))))...)))...... ( -34.00) >DroSec_CAF1 548 100 + 1 AUGCUAAU---CGAAAGUAAUUAACUUAA----------UGGCUUUGUCGAAGACCGUUUUGCGUGUAUAAUAGUGCGCCGAUUGGUUCAAAACGAAGCCCAAAGCAAUUUUU .((((...---...((((.....))))..----------.((((((((....(((((..(((.((((((....))))))))).)))))....))))))))...))))...... ( -27.80) >DroSim_CAF1 540 109 + 1 AUGCUAAU---CGAAAGUAAUUAACUUAAAUAUCGA-AAGGGCUUUGUCGAAGACCGUUUUGCGUGUAUAAUAGUGCGCCGAUUGGUUCAAAACGAAGCCCAAAGCAAUUUUU .((((..(---(((((((.....)))).....))))-..(((((((((....(((((..(((.((((((....))))))))).)))))....)))))))))..))))...... ( -34.50) >DroEre_CAF1 725 113 + 1 AGGAUAAAUAUUGAAUGUUAUCAUCUUAAAUAUCCAAAAGGGCUUUGUCGAAGACCGUUUUGCGUGUAGAAUAGUGCGCCGAUUGGUUCAAAACGAAGCCCAAAGCAAUUUUU .(((((..((.(((......)))...))..)))))....(((((((((....(((((.((.(((..(......)..))).)).)))))....)))))))))............ ( -32.40) >DroYak_CAF1 750 113 + 1 AGGAUAAAUAUUGAGUGUUAUCAACAUAAAUAUCAAUAAGGGCUUUGUCGAAGACCGUUUUGCGUGUAGAAUAGUGCGCCGAUUGGUUCAAAACGAAGCCCAAAGCAAUUUUU ........((((((...((((....))))...)))))).(((((((((....(((((.((.(((..(......)..))).)).)))))....)))))))))............ ( -32.20) >DroAna_CAF1 429 93 + 1 -----------GGAGAGUA--------CAGUGUAUA-GUGGGCUUUGUCGAAGACCGAUCUACGCUCAUAACAAUGUGUUUUUUGGUUCAAAACGAGGCCCAAAGCAAAUUUU -----------........--------...(((...-.((((((((((....((((((...((((..........))))...))))))....))))))))))..)))...... ( -26.90) >consensus AUGCUAAU___CGAAAGUAAUUAACUUAAAUAUCGA_AAGGGCUUUGUCGAAGACCGUUUUGCGUGUAGAAUAGUGCGCCGAUUGGUUCAAAACGAAGCCCAAAGCAAUUUUU .......................................(((((((((....(((((.((.(((..(......)..))).)).)))))....)))))))))............ (-24.14 = -24.25 + 0.11)

| Location | 13,016,676 – 13,016,788 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 78.99 |
| Mean single sequence MFE | -22.43 |
| Consensus MFE | -14.74 |
| Energy contribution | -14.97 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.688424 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
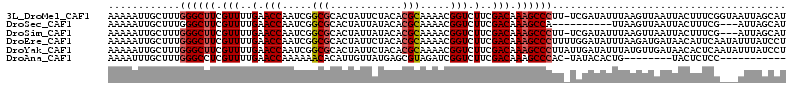
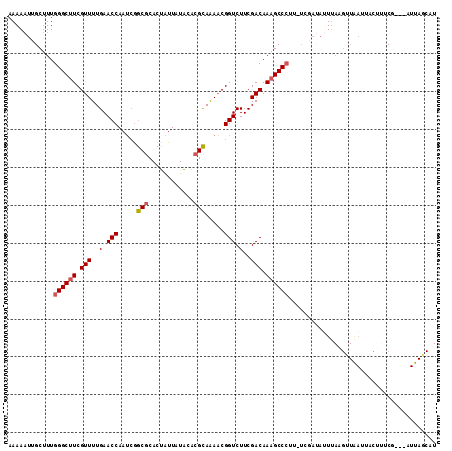
>3L_DroMel_CAF1 13016676 112 - 23771897 AAAAAUUGCUUUGGGCUUCGUUUUGAACCAAUCGGCGCACUAUUCUACACGCAAAACGGUCUUCGACAAAGCCCUU-UCGAUAUUUAAGUUAAUUACUUUCGGUAAUUAGCAU ....((((....((((((.(((..(.(((.....(((............))).....))).)..))).))))))..-.))))......(((((((((.....))))))))).. ( -27.30) >DroSec_CAF1 548 100 - 1 AAAAAUUGCUUUGGGCUUCGUUUUGAACCAAUCGGCGCACUAUUAUACACGCAAAACGGUCUUCGACAAAGCCA----------UUAAGUUAAUUACUUUCG---AUUAGCAU .............(((((.(((..(.(((.....(((............))).....))).)..))).))))).----------....(((((((......)---)))))).. ( -19.00) >DroSim_CAF1 540 109 - 1 AAAAAUUGCUUUGGGCUUCGUUUUGAACCAAUCGGCGCACUAUUAUACACGCAAAACGGUCUUCGACAAAGCCCUU-UCGAUAUUUAAGUUAAUUACUUUCG---AUUAGCAU ......((((..((((((.(((..(.(((.....(((............))).....))).)..))).))))))..-((((.....((((.....)))))))---)..)))). ( -23.30) >DroEre_CAF1 725 113 - 1 AAAAAUUGCUUUGGGCUUCGUUUUGAACCAAUCGGCGCACUAUUCUACACGCAAAACGGUCUUCGACAAAGCCCUUUUGGAUAUUUAAGAUGAUAACAUUCAAUAUUUAUCCU ............((((((.(((..(.(((.....(((............))).....))).)..))).))))))....(((((..((...(((......))).))..))))). ( -23.40) >DroYak_CAF1 750 113 - 1 AAAAAUUGCUUUGGGCUUCGUUUUGAACCAAUCGGCGCACUAUUCUACACGCAAAACGGUCUUCGACAAAGCCCUUAUUGAUAUUUAUGUUGAUAACACUCAAUAUUUAUCCU ............((((((.(((..(.(((.....(((............))).....))).)..))).)))))).....((((...(((((((......))))))).)))).. ( -24.00) >DroAna_CAF1 429 93 - 1 AAAAUUUGCUUUGGGCCUCGUUUUGAACCAAAAAACACAUUGUUAUGAGCGUAGAUCGGUCUUCGACAAAGCCCAC-UAUACACUG--------UACUCUCC----------- ...........(((((...(((((.......)))))...(((((..((.((.....)).))...))))).))))).-.........--------........----------- ( -17.60) >consensus AAAAAUUGCUUUGGGCUUCGUUUUGAACCAAUCGGCGCACUAUUAUACACGCAAAACGGUCUUCGACAAAGCCCUU_UCGAUAUUUAAGUUAAUUACUUUCG___AUUAGCAU ............((((((.(((..(.(((.....(((............))).....))).)..))).))))))....................................... (-14.74 = -14.97 + 0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:25:35 2006