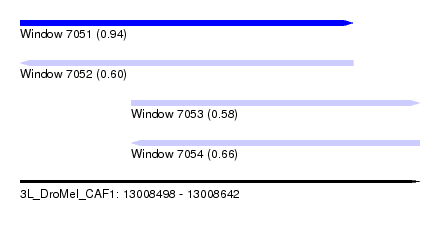
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,008,498 – 13,008,642 |
| Length | 144 |
| Max. P | 0.939520 |

| Location | 13,008,498 – 13,008,618 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.83 |
| Mean single sequence MFE | -37.50 |
| Consensus MFE | -35.62 |
| Energy contribution | -35.88 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939520 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13008498 120 + 23771897 UAUAGCUGGGCCAUGGUCAGGGUCGUCGGUUCAACGACAGGCCAUAAAACACGUUACAACUCUUACGGAACUUCAGUUCAGUAAGACUAGUUGUAUUUCCAGCAUGAGACAGCACUUCCC ....((((((..((((((...(((((.......))))).)))))).........((((((((((((.((((....)))).))))))...))))))..))))))................. ( -38.20) >DroSec_CAF1 15452 120 + 1 UAUAGCUGGGCCAUGGUCAGGGUCGUCGGUUCAACGACAGGCCAUAAAACACGUUACAACUCUUACGGAACUUCAGUUCAGUAAGACUAGUUGUAUUUUCAGCAUGAGACAGCACUUCCC ....((((..(((((.....((((((((......)))).))))...........((((((((((((.((((....)))).))))))...)))))).......)))).).))))....... ( -37.80) >DroSim_CAF1 19603 120 + 1 UAUAGCUGGGCCAUGGUCAGGGUCGUCGGUUCAACGACAGGCCAUAAAACACGUUACAACUCUUACGGAACUUCAGUUCAGUAAGACUAGUUGUAUUUUCAGCAUGAGACAGCACUUCCC ....((((..(((((.....((((((((......)))).))))...........((((((((((((.((((....)))).))))))...)))))).......)))).).))))....... ( -37.80) >DroYak_CAF1 17221 120 + 1 UGUAGCUGGGCCAUGGUCAGGGUCGUCGGUUCAACGACAGGCCAUAAAACACGUUACAACUCUUACGGAACCCCAGUUCAGUAAGACUAGUUGUACUUUGUGCAUGAGGCAGAACUUCUC .........(((((((((...(((((.......))))).))))))....((((.((((((((((((.((((....)))).))))))...))))))...)))).....))).......... ( -36.20) >consensus UAUAGCUGGGCCAUGGUCAGGGUCGUCGGUUCAACGACAGGCCAUAAAACACGUUACAACUCUUACGGAACUUCAGUUCAGUAAGACUAGUUGUAUUUUCAGCAUGAGACAGCACUUCCC ....((((...((((.....((((((((......)))).))))...........((((((((((((.((((....)))).))))))...)))))).......))))...))))....... (-35.62 = -35.88 + 0.25)

| Location | 13,008,498 – 13,008,618 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.83 |
| Mean single sequence MFE | -37.20 |
| Consensus MFE | -33.24 |
| Energy contribution | -33.42 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.601438 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13008498 120 - 23771897 GGGAAGUGCUGUCUCAUGCUGGAAAUACAACUAGUCUUACUGAACUGAAGUUCCGUAAGAGUUGUAACGUGUUUUAUGGCCUGUCGUUGAACCGACGACCCUGACCAUGGCCCAGCUAUA ((((.......))))..(((((...((((((...((((((.((((....)))).)))))))))))).......((((((.(.(((((((...)))))))...).)))))).))))).... ( -38.70) >DroSec_CAF1 15452 120 - 1 GGGAAGUGCUGUCUCAUGCUGAAAAUACAACUAGUCUUACUGAACUGAAGUUCCGUAAGAGUUGUAACGUGUUUUAUGGCCUGUCGUUGAACCGACGACCCUGACCAUGGCCCAGCUAUA (((..(((..(((....(((((((((((.((.(.((((((.((((....)))).)))))).).))...))))))).))))..(((((((...)))))))...))))))..)))....... ( -36.50) >DroSim_CAF1 19603 120 - 1 GGGAAGUGCUGUCUCAUGCUGAAAAUACAACUAGUCUUACUGAACUGAAGUUCCGUAAGAGUUGUAACGUGUUUUAUGGCCUGUCGUUGAACCGACGACCCUGACCAUGGCCCAGCUAUA (((..(((..(((....(((((((((((.((.(.((((((.((((....)))).)))))).).))...))))))).))))..(((((((...)))))))...))))))..)))....... ( -36.50) >DroYak_CAF1 17221 120 - 1 GAGAAGUUCUGCCUCAUGCACAAAGUACAACUAGUCUUACUGAACUGGGGUUCCGUAAGAGUUGUAACGUGUUUUAUGGCCUGUCGUUGAACCGACGACCCUGACCAUGGCCCAGCUACA ..(.((((..(((....((((....((((((...((((((.((((....)))).))))))))))))..))))...((((.(.(((((((...)))))))...).)))))))..)))).). ( -37.10) >consensus GGGAAGUGCUGUCUCAUGCUGAAAAUACAACUAGUCUUACUGAACUGAAGUUCCGUAAGAGUUGUAACGUGUUUUAUGGCCUGUCGUUGAACCGACGACCCUGACCAUGGCCCAGCUAUA ((((.......))))..(((((((((((.((.(.((((((.((((....)))).)))))).).))...)))))))..((((.(((((((...))))))).........)))))))).... (-33.24 = -33.42 + 0.19)

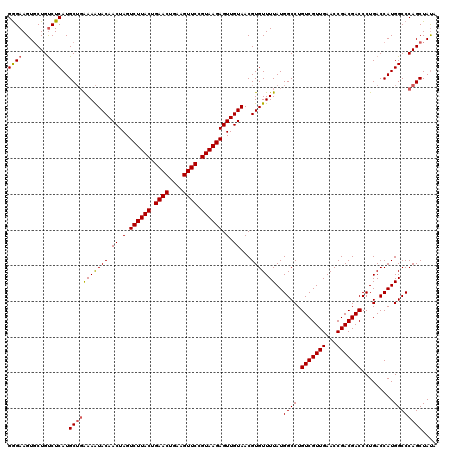
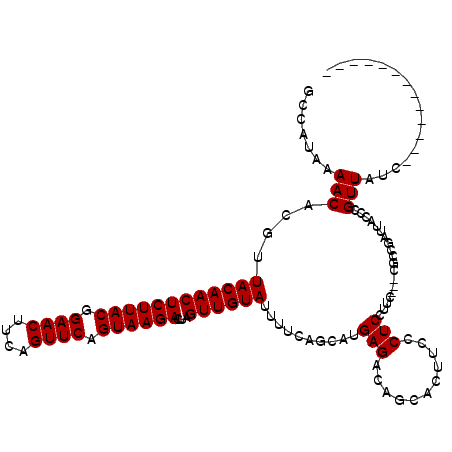
| Location | 13,008,538 – 13,008,642 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.71 |
| Mean single sequence MFE | -22.52 |
| Consensus MFE | -16.33 |
| Energy contribution | -16.33 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.576077 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13008538 104 + 23771897 GCCAUAAAACACGUUACAACUCUUACGGAACUUCAGUUCAGUAAGACUAGUUGUAUUUCCAGCAUGAGACAGCACUUCCCUCCUUC---CGCGGAUUACCCGUUAUC------------- ((............((((((((((((.((((....)))).))))))...))))))((((......))))..)).............---.((((.....))))....------------- ( -20.90) >DroSec_CAF1 15492 104 + 1 GCCAUAAAACACGUUACAACUCUUACGGAACUUCAGUUCAGUAAGACUAGUUGUAUUUUCAGCAUGAGACAGCACUUCCCUCCUUC---CGCCGAUUACCCGUUAUC------------- .......(((....((((((((((((.((((....)))).))))))...))))))...((.((..(((..............))).---.)).))......)))...------------- ( -18.04) >DroSim_CAF1 19643 103 + 1 GCCAUAAAACACGUUACAACUCUUACGGAACUUCAGUUCAGUAAGACUAGUUGUAUUUUCAGCAUGAGACAGCACUUCCCUCC-UC---CGCCGAUUACCCGUUAUC------------- .......(((....((((((((((((.((((....)))).))))))...))))))...((.((..(((..............)-))---.)).))......)))...------------- ( -19.04) >DroYak_CAF1 17261 120 + 1 GCCAUAAAACACGUUACAACUCUUACGGAACCCCAGUUCAGUAAGACUAGUUGUACUUUGUGCAUGAGGCAGAACUUCUCUCCUCCUAACUUCUCUUACCCGUUAUCAGCACGGGCCUAA (((...........((((((((((((.((((....)))).))))))...))))))...(((((..((((.(((.....))))))).((((...........))))...)))))))).... ( -32.10) >consensus GCCAUAAAACACGUUACAACUCUUACGGAACUUCAGUUCAGUAAGACUAGUUGUAUUUUCAGCAUGAGACAGCACUUCCCUCCUUC___CGCCGAUUACCCGUUAUC_____________ .......(((....((((((((((((.((((....)))).))))))...))))))..........(((...........)))...................)))................ (-16.33 = -16.33 + -0.00)

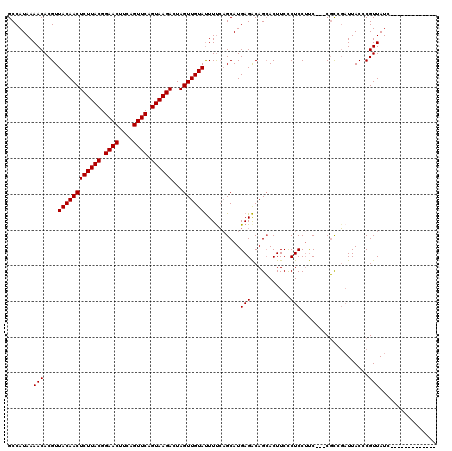
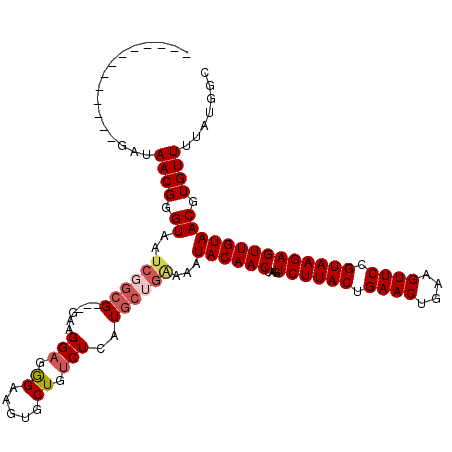
| Location | 13,008,538 – 13,008,642 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.71 |
| Mean single sequence MFE | -31.98 |
| Consensus MFE | -21.69 |
| Energy contribution | -23.12 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.661424 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13008538 104 - 23771897 -------------GAUAACGGGUAAUCCGCG---GAAGGAGGGAAGUGCUGUCUCAUGCUGGAAAUACAACUAGUCUUACUGAACUGAAGUUCCGUAAGAGUUGUAACGUGUUUUAUGGC -------------...((((.((..((((((---...(((.((.....)).)))..))).)))..((((((...((((((.((((....)))).)))))))))))))).))))....... ( -28.90) >DroSec_CAF1 15492 104 - 1 -------------GAUAACGGGUAAUCGGCG---GAAGGAGGGAAGUGCUGUCUCAUGCUGAAAAUACAACUAGUCUUACUGAACUGAAGUUCCGUAAGAGUUGUAACGUGUUUUAUGGC -------------...((((.((..((((((---...(((.((.....)).)))..))))))...((((((...((((((.((((....)))).)))))))))))))).))))....... ( -30.50) >DroSim_CAF1 19643 103 - 1 -------------GAUAACGGGUAAUCGGCG---GA-GGAGGGAAGUGCUGUCUCAUGCUGAAAAUACAACUAGUCUUACUGAACUGAAGUUCCGUAAGAGUUGUAACGUGUUUUAUGGC -------------...((((.((..((((((---..-(((.((.....)).)))..))))))...((((((...((((((.((((....)))).)))))))))))))).))))....... ( -32.00) >DroYak_CAF1 17261 120 - 1 UUAGGCCCGUGCUGAUAACGGGUAAGAGAAGUUAGGAGGAGAGAAGUUCUGCCUCAUGCACAAAGUACAACUAGUCUUACUGAACUGGGGUUCCGUAAGAGUUGUAACGUGUUUUAUGGC ....((((((.......)))))).......((...((((((((...)))).))))..))(((...((((((...((((((.((((....)))).))))))))))))...)))........ ( -36.50) >consensus _____________GAUAACGGGUAAUCGGCG___GAAGGAGGGAAGUGCUGUCUCAUGCUGAAAAUACAACUAGUCUUACUGAACUGAAGUUCCGUAAGAGUUGUAACGUGUUUUAUGGC ................((((.((..((((((......(((.((.....)).)))..))))))...((((((...((((((.((((....)))).)))))))))))))).))))....... (-21.69 = -23.12 + 1.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:25:28 2006