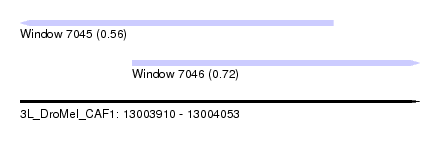
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,003,910 – 13,004,053 |
| Length | 143 |
| Max. P | 0.722674 |

| Location | 13,003,910 – 13,004,022 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.27 |
| Mean single sequence MFE | -32.62 |
| Consensus MFE | -19.48 |
| Energy contribution | -19.64 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.559597 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
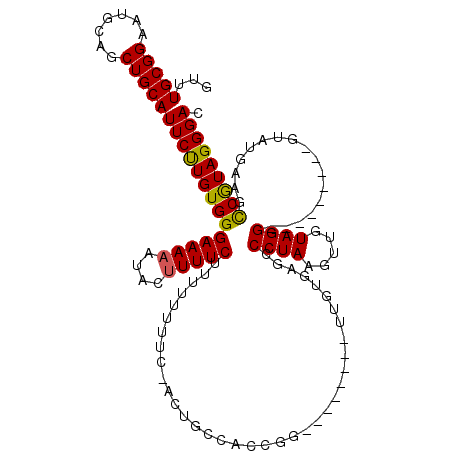
>3L_DroMel_CAF1 13003910 112 - 23771897 GCUCACAA--------CCGGUGGCAGUAGAAAAAAAAGAAAAGUAUUUUUCCCACAAGAAUGCAGCUGCAUUCCGCAAACCACCACCAUGCCCAAUCGCUGGGUCAGAUCUCCUGCCGCA ........--------...(((((((.(((...........................((((((....))))))................(((((.....)))))....))).))))))). ( -28.90) >DroSec_CAF1 10952 111 - 1 GAUCACAA--------CCGGUGGCGGUGGAAA-AAAAGAAAUGUAUUUUUCCCACAAGAAUGCAGCUGCAUUCCGCAAACCACCACCAUGCCCAAUCGCUGGGUCAGAUCUCCUGCCGCA ((((....--------..(((((.((((((((-((.(....)...))))))).....((((((....)))))).....))).)))))..(((((.....)))))..)))).......... ( -36.70) >DroSim_CAF1 13480 112 - 1 GAUCACAA--------CCGGUGGCGGUGGAAAAAAAAGAAAAGUAUUUUUCCCACAAGAAUGCAGCUGCAUUCCGCAAACCACCACCAUGCCCAAUCGCUGGGUCAGAUCUCCUGCCGCA ((((....--------..(((((.((((((((((...........))))))).....((((((....)))))).....))).)))))..(((((.....)))))..)))).......... ( -35.90) >DroEre_CAF1 10861 112 - 1 GCUCACAACUGCUAUAGAGCUGCUAAU-----AGAGAGAAAAGUCUUUUUCCCACAGGAAUGCAGCUGCAUUCCGCA---CACCAGCGUGCCCAAUCGCAGGGUCAGAUCUGCUGCCGCA ((......((((.((.(.((((((...-----(((((((....)))))))......(((((((....)))))))...---....)))).)).).)).))))((.(((.....))))))). ( -33.10) >DroYak_CAF1 11369 104 - 1 GCUCACAA--------GCGGUGGUAGU-----AAAAAGAAAAGUAUUUUUCCCACAGGAAUGCAGCUGCAGUCCGCA---CAACACCGUGUCCAAUCGCAGGGUCAGAUCUCCUGCCGCA ((......--------((((((((.(.-----(((((........))))).).)).(((.(((....))).)))...---...))))))........((((((.......)))))).)). ( -28.50) >consensus GCUCACAA________CCGGUGGCAGU_GAAAAAAAAGAAAAGUAUUUUUCCCACAAGAAUGCAGCUGCAUUCCGCAAACCACCACCAUGCCCAAUCGCUGGGUCAGAUCUCCUGCCGCA ...................(((((((...............................((((((....))))))...............(((((.......))).))......))))))). (-19.48 = -19.64 + 0.16)


| Location | 13,003,950 – 13,004,053 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.72 |
| Mean single sequence MFE | -33.46 |
| Consensus MFE | -16.22 |
| Energy contribution | -15.86 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.722674 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13003950 103 + 23771897 GUUUGCGGAAUGCAGCUGCAUUCUUGUGGGAAAAAUACUUUUCUUUUUUUUCUACUGCCACCGG--------UUGUGAGCCCUAAAUUGUAGG---------GUAUGAAGCCGUAGGGAC ......(((((((....))))))).((((((((((..........))))))))))..((..(((--------((....((((((.....))))---------))....)))))..))... ( -33.10) >DroSec_CAF1 10992 102 + 1 GUUUGCGGAAUGCAGCUGCAUUCUUGUGGGAAAAAUACAUUUCUUUU-UUUCCACCGCCACCGG--------UUGUGAUCCCUAAAUUGUAGG---------GUAUGAAGCCGUAGGGAU ......(((((((....))))))).((((((((((.........)))-)))))))..((..(((--------((.....(((((.....))))---------).....)))))..))... ( -33.80) >DroSim_CAF1 13520 103 + 1 GUUUGCGGAAUGCAGCUGCAUUCUUGUGGGAAAAAUACUUUUCUUUUUUUUCCACCGCCACCGG--------UUGUGAUCCCUAAGUUGUAGG---------GUAUGAAGCCGUAGGGAC ......(((((((....))))))).((((((((((..........))))))))))..((..(((--------((.....(((((.....))))---------).....)))))..))... ( -33.70) >DroEre_CAF1 10901 103 + 1 ---UGCGGAAUGCAGCUGCAUUCCUGUGGGAAAAAGACUUUUCUCUCU-----AUUAGCAGCUCUAUAGCAGUUGUGAGCCCUAAGUUGUAGG---------GUAUGAAAUCGUAGGGAC ---((((((((((....))))))).(.(((((((....))))))).).-----....))).((((((.((....))..((((((.....))))---------))........)))))).. ( -31.60) >DroYak_CAF1 11409 104 + 1 ---UGCGGACUGCAGCUGCAUUCCUGUGGGAAAAAUACUUUUCUUUUU-----ACUACCACCGC--------UUGUGAGCCCUAAGAUGUAGGGUAUGUAGGGUAUGAGCCCAUAGGGAC ---(((((.(....))))))((((((((((....(((((((.(.....-----.....(((...--------..))).((((((.....))))))..).)))))))...)))))))))). ( -35.10) >consensus GUUUGCGGAAUGCAGCUGCAUUCUUGUGGGAAAAAUACUUUUCUUUUUUUUC_ACUGCCACCGG________UUGUGAGCCCUAAGUUGUAGG_________GUAUGAAGCCGUAGGGAC ...(((((.......)))))((((((((((((((....))))).....................................((((.....)))).................))))))))). (-16.22 = -15.86 + -0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:25:21 2006