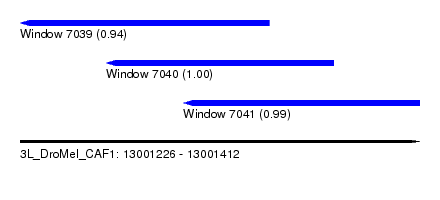
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,001,226 – 13,001,412 |
| Length | 186 |
| Max. P | 0.996534 |

| Location | 13,001,226 – 13,001,342 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.30 |
| Mean single sequence MFE | -28.33 |
| Consensus MFE | -28.28 |
| Energy contribution | -28.28 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.45 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937248 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13001226 116 - 23771897 UAUCGAAAAAUGAAGAAAACGCCGAAAGGCCACAGAAACUUUCAUAGUGGUCG----UAACUUUUUCCCCAUCGGCCAAACGACAUUUGAAGUCAAUUGUCGUCGCGUCGUGCAGAAAAA ...(((...((((((.....(((....))).........)))))).....)))----.....(((((..((.((((...((((((.(((....))).))))))...))))))..))))). ( -28.14) >DroSec_CAF1 8260 116 - 1 UAUCGAAAAAUGAAGAAAACGCCGAAAGGCAACAGAAACUUUCAUAGUGGUCG----UAACUUUUUCCCCAUCGGCCAAACGACAUUUGAAGUCAAUUGUCGUCGCGUCGUGCAGAAAAA ..............((..(((((....)))....((.....))...))..)).----.....(((((..((.((((...((((((.(((....))).))))))...))))))..))))). ( -29.00) >DroSim_CAF1 10821 116 - 1 UAUCGAAAAAUGAAGAAAACGCCGAAAGGCCACAGAAACUUUCAUAGUGGUCG----UAACUUUUUCCCCAUCGGCCAAACGACAUUUGAAGUCAAUUGUCGUCGCGUCGUGCAGAAAAA ...(((...((((((.....(((....))).........)))))).....)))----.....(((((..((.((((...((((((.(((....))).))))))...))))))..))))). ( -28.14) >DroEre_CAF1 8044 116 - 1 UAUCGAAAAAUGAAGAAAACGCCGAAAGGCCACAGAAACUUUCAUAGUGGUCG----UAACUUUUUCCCCAUCGGCCAAACGACAUUUGAAGUCAAUUGUCGUCGCGUCGUGCAGAAAAA ...(((...((((((.....(((....))).........)))))).....)))----.....(((((..((.((((...((((((.(((....))).))))))...))))))..))))). ( -28.14) >DroYak_CAF1 8438 120 - 1 UAUCGAAAAAUGAAGAAAACGCCGAAAGGCCACAGAAACUUUCAUAGUGGUCGUAUGUAACUUUUUCCCCAUCGGCCAAACGACAUUUGAAGUCAAUUGUCGUCGCGUCGUGCAGAAAAA ((((((...((((((.....(((....))).........)))))).....))).))).....(((((..((.((((...((((((.(((....))).))))))...))))))..))))). ( -28.24) >consensus UAUCGAAAAAUGAAGAAAACGCCGAAAGGCCACAGAAACUUUCAUAGUGGUCG____UAACUUUUUCCCCAUCGGCCAAACGACAUUUGAAGUCAAUUGUCGUCGCGUCGUGCAGAAAAA ..............((..(((((....)))....((.....))...))..))..........(((((..((.((((...((((((.(((....))).))))))...))))))..))))). (-28.28 = -28.28 + -0.00)

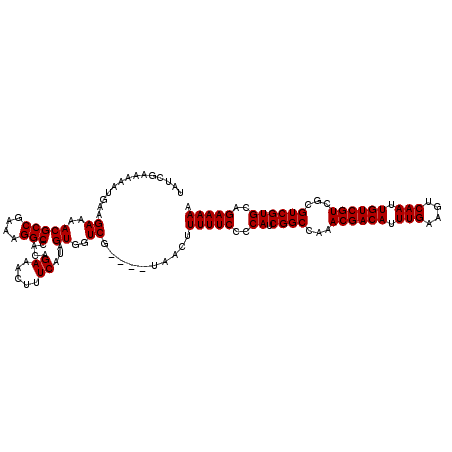
| Location | 13,001,266 – 13,001,372 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.98 |
| Mean single sequence MFE | -27.55 |
| Consensus MFE | -23.07 |
| Energy contribution | -23.67 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.71 |
| SVM RNA-class probability | 0.996534 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13001266 106 - 23771897 GCUAU------A----UUCGUUUCAAUUCGUUUCGGUUAUUAUCGAAAAAUGAAGAAAACGCCGAAAGGCCACAGAAACUUUCAUAGUGGUCG----UAACUUUUUCCCCAUCGGCCAAA (((((------.----...(((((..(((((((((((....)))))..))))))......(((....)))....)))))....)))))(((((----...............)))))... ( -27.96) >DroSec_CAF1 8300 106 - 1 GCUAU------A----UUCGUUUCAAUUCGUUUCGGUUAUUAUCGAAAAAUGAAGAAAACGCCGAAAGGCAACAGAAACUUUCAUAGUGGUCG----UAACUUUUUCCCCAUCGGCCAAA (((((------.----...(((((..(((((((((((....)))))..))))))......(((....)))....)))))....)))))(((((----...............)))))... ( -28.86) >DroSim_CAF1 10861 106 - 1 GCUAU------A----UUCGUUUCAAUUCGUUUCGGUUAUUAUCGAAAAAUGAAGAAAACGCCGAAAGGCCACAGAAACUUUCAUAGUGGUCG----UAACUUUUUCCCCAUCGGCCAAA (((((------.----...(((((..(((((((((((....)))))..))))))......(((....)))....)))))....)))))(((((----...............)))))... ( -27.96) >DroEre_CAF1 8084 112 - 1 GCUAUGUAUAUA----UUCGUUACAAUUCGCUUCGGUUAUUAUCGAAAAAUGAAGAAAACGCCGAAAGGCCACAGAAACUUUCAUAGUGGUCG----UAACUUUUUCCCCAUCGGCCAAA ((((((......----((((((....((((.((((((....))))))...))))...)))(((....)))....))).....))))))(((((----...............)))))... ( -26.66) >DroYak_CAF1 8478 120 - 1 GCCAUAUAUAUAUACAUACGUUUCAAUUCGCUUCGGUUAUUAUCGAAAAAUGAAGAAAACGCCGAAAGGCCACAGAAACUUUCAUAGUGGUCGUAUGUAACUUUUUCCCCAUCGGCCAAA (((((....)).(((((((((((...((((.((((((....))))))...))))..))))(((....)))....((.(((.....)))..)))))))))..............))).... ( -26.30) >consensus GCUAU______A____UUCGUUUCAAUUCGUUUCGGUUAUUAUCGAAAAAUGAAGAAAACGCCGAAAGGCCACAGAAACUUUCAUAGUGGUCG____UAACUUUUUCCCCAUCGGCCAAA ...................(((((..(((((((((((....))))))..)))))......(((....)))....)))))........((((((...................)))))).. (-23.07 = -23.67 + 0.60)

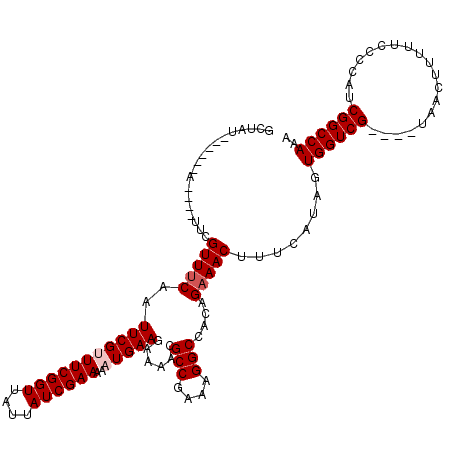
| Location | 13,001,302 – 13,001,412 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.18 |
| Mean single sequence MFE | -25.52 |
| Consensus MFE | -23.90 |
| Energy contribution | -24.54 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.37 |
| SVM RNA-class probability | 0.992999 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13001302 110 - 23771897 UUAAAAGGCGCUUUUCAUUUUCUUUUGGGAAAGUUAUCAAGCUAU------A----UUCGUUUCAAUUCGUUUCGGUUAUUAUCGAAAAAUGAAGAAAACGCCGAAAGGCCACAGAAACU .........((((...(((((((....)))))))....))))...------.----...(((((..(((((((((((....)))))..))))))......(((....)))....))))). ( -26.40) >DroSec_CAF1 8336 110 - 1 UUAAAAGGCGCUUUUCAUUUUCUUUUGGGAAAGUUAUCAAGCUAU------A----UUCGUUUCAAUUCGUUUCGGUUAUUAUCGAAAAAUGAAGAAAACGCCGAAAGGCAACAGAAACU .........((((...(((((((....)))))))....))))...------.----...(((((..(((((((((((....)))))..))))))......(((....)))....))))). ( -27.30) >DroSim_CAF1 10897 110 - 1 UUAAAAGGCGCUUUUCAUUUUCUUUUGGGAAAGUUAUCAAGCUAU------A----UUCGUUUCAAUUCGUUUCGGUUAUUAUCGAAAAAUGAAGAAAACGCCGAAAGGCCACAGAAACU .........((((...(((((((....)))))))....))))...------.----...(((((..(((((((((((....)))))..))))))......(((....)))....))))). ( -26.40) >DroEre_CAF1 8120 116 - 1 UUAAAAGGCGCUUUUCAUUUUCUUUUGGGAAAGUUAUCAAGCUAUGUAUAUA----UUCGUUACAAUUCGCUUCGGUUAUUAUCGAAAAAUGAAGAAAACGCCGAAAGGCCACAGAAACU .........((((...(((((((....)))))))....))))..........----((((((....((((.((((((....))))))...))))...)))(((....)))....)))... ( -22.20) >DroYak_CAF1 8518 120 - 1 UUAAAAGGCGCUUUUCAUUUGCUUUUGGGAAAGUUAUCAAGCCAUAUAUAUAUACAUACGUUUCAAUUCGCUUCGGUUAUUAUCGAAAAAUGAAGAAAACGCCGAAAGGCCACAGAAACU ......((((((((((...........)))))))......)))................(((((..((((.((((((....))))))...))))......(((....)))....))))). ( -25.30) >consensus UUAAAAGGCGCUUUUCAUUUUCUUUUGGGAAAGUUAUCAAGCUAU______A____UUCGUUUCAAUUCGUUUCGGUUAUUAUCGAAAAAUGAAGAAAACGCCGAAAGGCCACAGAAACU ......(((.......(((((((....)))))))......)))................(((((..(((((((((((....))))))..)))))......(((....)))....))))). (-23.90 = -24.54 + 0.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:25:16 2006