
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,997,710 – 12,997,863 |
| Length | 153 |
| Max. P | 0.653442 |

| Location | 12,997,710 – 12,997,830 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.14 |
| Mean single sequence MFE | -38.03 |
| Consensus MFE | -30.06 |
| Energy contribution | -29.78 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.79 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12997710 120 + 23771897 GCGCCAUGGACAUUAACACUGCCCUCCAGGAGGUCUUGAAGAAGUCCCUGAUCGCCGAUGGACUCGUCCAUGGCAUCCACCAGGCCUGCAAGGCCCUGGACAAGUAAGUAGUCCCACAAU ..(((((((((.((((.(((..((....)).))).)))).((.((((.((.....))..))))))))))))))).....(((((((.....)).)))))..................... ( -39.90) >DroVir_CAF1 4354 120 + 1 GCGCCAUGGACAUAAACACCGCGCUACAGGAAGUUCUAAAGAAGUCGCUGAUCGCUGAUGGACUCGUCCAUGGCAUCCACCAGGCCUGCAAGGCUUUGGACAAGUAAGCAACAACAUCAU (((((((((((.....(((.(((...(((((..(((....))).)).)))..))).).)).....))))))))).((((..(((((.....))))))))).......))........... ( -39.50) >DroGri_CAF1 4225 120 + 1 GCGCCAUGGACAUAAACACCGCGCUGCAGGAAGUGCUGAAGAAGUCGCUGAUCGCCGAUGGCCUCGUCCAUGGCAUCCACCAAGCCUGCAAAGCUCUGGACAAGUAAGCAAAAAAUUCAA (((((((((((.........(((((......))))).((.(..(((((.....).))))..).))))))))))).....((((((.......))).)))........))........... ( -38.20) >DroWil_CAF1 6281 119 + 1 GUGCCAUGGACAUUAACACCGCUCUCCAGGAAGUUCUGAAGAAGUCUCUGAUUGCCGAUGGUUUGGUCCAUGGCAUCCAUCAGGCUUGCAAGGCUUUGGACAAGUAAGUUUCCC-UCUCU .((((((((((...(((.(.((....(((..(.(((....))).)..)))...)).)...)))..))))))))))((((..(((((.....)))))))))..............-..... ( -36.20) >DroMoj_CAF1 4202 116 + 1 GCGCCAUGGACAUAAACACCGCGCUACAGGAAGUGCUGAAGAAGUCGCUGAUCGCCGAUGGACUCGUCCAUGGCAUCCACCAGGCCUGCAAGGCAUUGGACAAGUAAGCAAAAACA---- (((((((((((.........(((((......))))).((....(((((.....).))))....))))))))))).....(((((((.....))).))))........)).......---- ( -39.50) >DroPer_CAF1 4279 117 + 1 GCGCCAUGGACAUUAACACUGCCCUGCAGGAAGUGCUGAAGAAGUCUCUGAUUGCUGAUGGUCUCGUGCAUGGCAUCCACCAGGCCUGCAAGGCCCUGGACAAGUAAGUAGACCAAG--- ..((((((.((.(((.((((..((....)).)))).))).((..((((........)).))..)))).)))))).....(((((((.....)).)))))..................--- ( -34.90) >consensus GCGCCAUGGACAUAAACACCGCGCUACAGGAAGUGCUGAAGAAGUCGCUGAUCGCCGAUGGACUCGUCCAUGGCAUCCACCAGGCCUGCAAGGCUCUGGACAAGUAAGCAAAAACAC_AU (((((((((((.....(((.((....(((((..(.......)..)).)))...)).).)).....))))))))).((((...((((.....)))).)))).......))........... (-30.06 = -29.78 + -0.27)

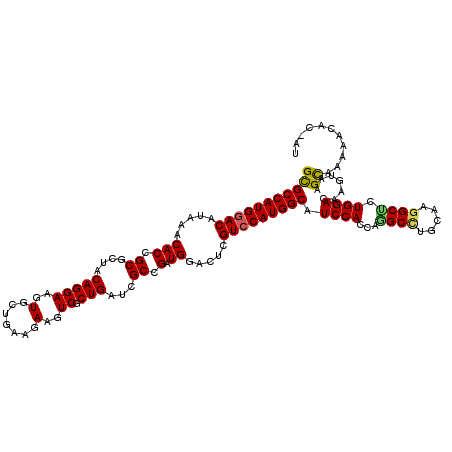

| Location | 12,997,750 – 12,997,863 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 83.32 |
| Mean single sequence MFE | -42.01 |
| Consensus MFE | -29.20 |
| Energy contribution | -30.56 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.653442 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12997750 113 - 23771897 AGAGCCCA---AGUGAAUAACCACGUUGACCGCCAGAUUGUGGGACUACUUACUUGUCCAGGGCCUUGCAGGCCUGGUGGAUGCCAUGGACGAGUCCAUCGGCGAUCAGGGACUUC .((((((.---.(((......)))..((((((((........((((.........)))).(((((.....)))))))))).(((((((((....))))).)))).)))))).))). ( -44.70) >DroSec_CAF1 4830 113 - 1 AGAGUCCA---AGUGAAUAACCACGUUGACCGCAAAAUUGUGGGACUACUUACUUGUCCAGGGCCUUGCAGGCCUGGUGGAUGCCAUGGACGAGUCCAUCGGCGAUAAGGGACUUC .((((((.---.(((......)))((((((((((....)))))........((((((((((((((.....)))))(((....))).)))))))))...)))))......)))))). ( -45.00) >DroEre_CAF1 4638 116 - 1 AAAUCCCAAGAAGUGAAUAAGCACCUUGAUGUCUGGAUUGUGCGACUACUUACUUGUCCAGGGCCUUGCAGGCCUGGUGGAUGCCAUGGACGAGUCCAUCGGCGAUCAGGGACUUC ...((((..............((((.......((((((.(((........)))..))))))((((.....)))).))))(((((((((((....))))).))).))).)))).... ( -41.40) >DroYak_CAF1 5078 116 - 1 AAACCCCAAGAAGUGUAUAACCACGUUGAUCUCCGGAUUGUGGGACUACUUACUUGUCCAGGGCCUUGCAGGCCUGGUGGAUGCCAUGGACGAGUCCAUCGGCGAUCAGGGACUUC ....(((.....(((......)))..((((((((((((.(((((....)))))..)))).(((((.....)))))...))).((((((((....))))).)))))))))))..... ( -44.10) >DroAna_CAF1 9625 104 - 1 -----CCGAACUAUAGAGAAGCAUCCGG-------GUCUCUGGGAGAACUUACUUGUCCAGAGCCUUGCAGGCCUGGUGGAUGCCAUGGACGAGGCCAUCGGCGAUCAGGGACUUC -----(((.................)))-------((((((((.........((((((((..(((.....))).((((....))))))))))))(((...)))..))))))))... ( -34.83) >consensus AAAGCCCAA__AGUGAAUAACCACGUUGACCGCCAGAUUGUGGGACUACUUACUUGUCCAGGGCCUUGCAGGCCUGGUGGAUGCCAUGGACGAGUCCAUCGGCGAUCAGGGACUUC ....(((.....(((......)))...........((((((.((.......((((((((((((((.....)))))(((....))).)))))))))...)).)))))).)))..... (-29.20 = -30.56 + 1.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:25:11 2006