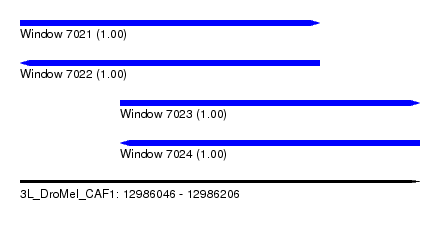
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,986,046 – 12,986,206 |
| Length | 160 |
| Max. P | 0.999974 |

| Location | 12,986,046 – 12,986,166 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.67 |
| Mean single sequence MFE | -28.19 |
| Consensus MFE | -26.62 |
| Energy contribution | -27.26 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.94 |
| SVM decision value | 4.88 |
| SVM RNA-class probability | 0.999958 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12986046 120 + 23771897 CCCAACUACAUUUUCAAAAAGAACAAGUUUCGCUUCUUUUUAACCUUUUCCAAAGCUGGUGCCUGUUCGGACUCAUCGACGUCCUUGUCGAGGUCCGACUUUUUGCCCUUGCCCAGCUUA ...............(((((((((.......).))))))))...........(((((((.((..(((((((((..((((((....)))))))))))))......))....))))))))). ( -32.20) >DroSec_CAF1 103372 120 + 1 UACUACUACAUUUUCAUAAAGAACAAGUUUCGCUUCUUUUUAACCUUUUCCAAAGCUGGUGCCUGUUCGGACUCAUCGACGUCCUUGUCGAGGUCCGACUUUUUGCCCUUGCCCAGCUUA ..................(((((.(((.....))).)))))...........(((((((.((..(((((((((..((((((....)))))))))))))......))....))))))))). ( -30.80) >DroSim_CAF1 113006 120 + 1 UCCAACUACAUUUUCAUAAAGAACAAGUUUCGCUUCUUUUUAACCUUUUCCAAAGCUGGUGCCUGUUCGGACUCAUCGACGUCCUUGUCGAGGUCCGACUUUUUGCCCUUGCCCAGCUUA ..................(((((.(((.....))).)))))...........(((((((.((..(((((((((..((((((....)))))))))))))......))....))))))))). ( -30.80) >DroEre_CAF1 109369 120 + 1 CCCUCCUACAUUUUCAAAAAGAACAAGUUUCGCUUCUUUUUAACCUUUUCCAAAGCUGGUGCCUGUUCAGACUCAUCGACGUCCUUAUCGAGGUCUGACUUUUUGCCCUUGCCCAACUUA ........((....(((((((.(((.((.((((((.................)))).)).)).)))(((((((..((((........))))))))))))))))))....))......... ( -20.43) >DroYak_CAF1 108156 120 + 1 CCCUCCUACAUUUUCAGAAAGAACAAGUUUCGCUUCUUUUUAACUUUUUCCAAAGCUGGUGCUUGUUCAGACUCAUCGACGUCCUUGUCGAGAUCUGACUUUUUGCCCUUGCCCAACUUA ........((....(((((((.((((((.((((((.................)))).)).))))))(((((.((.((((((....))))))))))))))))))))....))......... ( -26.73) >consensus CCCUACUACAUUUUCAAAAAGAACAAGUUUCGCUUCUUUUUAACCUUUUCCAAAGCUGGUGCCUGUUCGGACUCAUCGACGUCCUUGUCGAGGUCCGACUUUUUGCCCUUGCCCAGCUUA ...............(((((((((.......).))))))))...........(((((((.((..(((((((((..((((((....)))))))))))))......))....))))))))). (-26.62 = -27.26 + 0.64)


| Location | 12,986,046 – 12,986,166 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.67 |
| Mean single sequence MFE | -33.68 |
| Consensus MFE | -30.88 |
| Energy contribution | -29.92 |
| Covariance contribution | -0.96 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.92 |
| SVM decision value | 4.11 |
| SVM RNA-class probability | 0.999800 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12986046 120 - 23771897 UAAGCUGGGCAAGGGCAAAAAGUCGGACCUCGACAAGGACGUCGAUGAGUCCGAACAGGCACCAGCUUUGGAAAAGGUUAAAAAGAAGCGAAACUUGUUCUUUUUGAAAAUGUAGUUGGG .(((((((((...(........((((((((((((......))))).).)))))).)..)).))))))).........((((((((((.(((...)))))))))))))............. ( -35.30) >DroSec_CAF1 103372 120 - 1 UAAGCUGGGCAAGGGCAAAAAGUCGGACCUCGACAAGGACGUCGAUGAGUCCGAACAGGCACCAGCUUUGGAAAAGGUUAAAAAGAAGCGAAACUUGUUCUUUAUGAAAAUGUAGUAGUA ...((((.(((.((((......((((((((((((......))))).).))))))....)).))((((((....))))))..((((((.(((...))))))))).......)))..)))). ( -33.20) >DroSim_CAF1 113006 120 - 1 UAAGCUGGGCAAGGGCAAAAAGUCGGACCUCGACAAGGACGUCGAUGAGUCCGAACAGGCACCAGCUUUGGAAAAGGUUAAAAAGAAGCGAAACUUGUUCUUUAUGAAAAUGUAGUUGGA ..(((((((((((.((......((((((((((((......))))).).))))))....))...((((((....))))))..............)))))))..(((....))).))))... ( -32.80) >DroEre_CAF1 109369 120 - 1 UAAGUUGGGCAAGGGCAAAAAGUCAGACCUCGAUAAGGACGUCGAUGAGUCUGAACAGGCACCAGCUUUGGAAAAGGUUAAAAAGAAGCGAAACUUGUUCUUUUUGAAAAUGUAGGAGGG ((((((..((..((((......((((((((((((......))))).).))))))....)).))((((((....))))))........))..))))))..(((((((......))))))). ( -32.70) >DroYak_CAF1 108156 120 - 1 UAAGUUGGGCAAGGGCAAAAAGUCAGAUCUCGACAAGGACGUCGAUGAGUCUGAACAAGCACCAGCUUUGGAAAAAGUUAAAAAGAAGCGAAACUUGUUCUUUCUGAAAAUGUAGGAGGG ((((((..((..((((......((((((((((((......))))).)).)))))....)).))((((((....))))))........))..))))))..(((((((......))))))). ( -34.40) >consensus UAAGCUGGGCAAGGGCAAAAAGUCGGACCUCGACAAGGACGUCGAUGAGUCCGAACAGGCACCAGCUUUGGAAAAGGUUAAAAAGAAGCGAAACUUGUUCUUUAUGAAAAUGUAGUAGGG ......(((((((.((......((((((((((((......))))).).))))))....))...((((((....))))))..............))))))).................... (-30.88 = -29.92 + -0.96)



| Location | 12,986,086 – 12,986,206 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.58 |
| Mean single sequence MFE | -28.12 |
| Consensus MFE | -25.82 |
| Energy contribution | -25.90 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.92 |
| SVM decision value | 4.55 |
| SVM RNA-class probability | 0.999918 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12986086 120 + 23771897 UAACCUUUUCCAAAGCUGGUGCCUGUUCGGACUCAUCGACGUCCUUGUCGAGGUCCGACUUUUUGCCCUUGCCCAGCUUAUUUUCCUCCUUAAGUUUUUUUCGUUCCUUUUUAGACAGCU ............(((((((.((..(((((((((..((((((....)))))))))))))......))....))))))))).......................(((........))).... ( -28.80) >DroSec_CAF1 103412 120 + 1 UAACCUUUUCCAAAGCUGGUGCCUGUUCGGACUCAUCGACGUCCUUGUCGAGGUCCGACUUUUUGCCCUUGCCCAGCUUAUUUUCCUCCUUAAGUUUUUUUCGUUCCUUUUUGGACAGCU ............(((((((.((..(((((((((..((((((....)))))))))))))......))....))))))))).......................((.((.....)))).... ( -31.20) >DroSim_CAF1 113046 120 + 1 UAACCUUUUCCAAAGCUGGUGCCUGUUCGGACUCAUCGACGUCCUUGUCGAGGUCCGACUUUUUGCCCUUGCCCAGCUUAUUUUCCUCCUUAAGUUUUUUUCGUUCCUUUUUGGACAGCU ............(((((((.((..(((((((((..((((((....)))))))))))))......))....))))))))).......................((.((.....)))).... ( -31.20) >DroEre_CAF1 109409 120 + 1 UAACCUUUUCCAAAGCUGGUGCCUGUUCAGACUCAUCGACGUCCUUAUCGAGGUCUGACUUUUUGCCCUUGCCCAACUUAUUUUCCUCCUUAAGUUUUUUUCGCUCCUUUUUUGACAACU ..........(((((..((.((....(((((((..((((........)))))))))))................((((((..........))))))......)).))..)))))...... ( -21.80) >DroYak_CAF1 108196 120 + 1 UAACUUUUUCCAAAGCUGGUGCUUGUUCAGACUCAUCGACGUCCUUGUCGAGAUCUGACUUUUUGCCCUUGCCCAACUUAUUUUCCUCCUUAAGUUUUUUUCGCUCCUUUUUGGACAACU ........(((((((..((.((....(((((.((.((((((....)))))))))))))................((((((..........))))))......)).)).)))))))..... ( -27.60) >consensus UAACCUUUUCCAAAGCUGGUGCCUGUUCGGACUCAUCGACGUCCUUGUCGAGGUCCGACUUUUUGCCCUUGCCCAGCUUAUUUUCCUCCUUAAGUUUUUUUCGUUCCUUUUUGGACAGCU ........((((((((.((.((....(((((((..((((((....)))))))))))))......))))..))..((((((..........)))))).............))))))..... (-25.82 = -25.90 + 0.08)

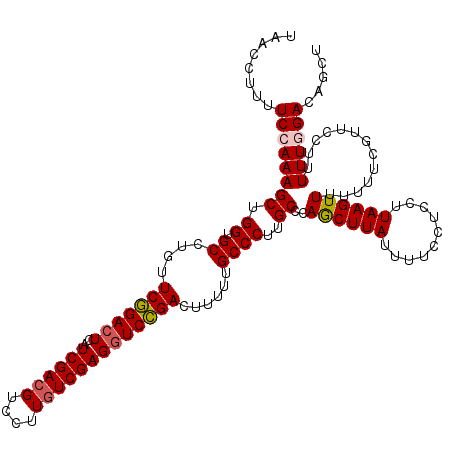
| Location | 12,986,086 – 12,986,206 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.58 |
| Mean single sequence MFE | -32.03 |
| Consensus MFE | -30.38 |
| Energy contribution | -29.02 |
| Covariance contribution | -1.36 |
| Combinations/Pair | 1.21 |
| Mean z-score | -3.06 |
| Structure conservation index | 0.95 |
| SVM decision value | 5.11 |
| SVM RNA-class probability | 0.999974 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12986086 120 - 23771897 AGCUGUCUAAAAAGGAACGAAAAAAACUUAAGGAGGAAAAUAAGCUGGGCAAGGGCAAAAAGUCGGACCUCGACAAGGACGUCGAUGAGUCCGAACAGGCACCAGCUUUGGAAAAGGUUA ((((.((((((...............(((....))).......(((((((...(........((((((((((((......))))).).)))))).)..)).)))))))))))...)))). ( -33.10) >DroSec_CAF1 103412 120 - 1 AGCUGUCCAAAAAGGAACGAAAAAAACUUAAGGAGGAAAAUAAGCUGGGCAAGGGCAAAAAGUCGGACCUCGACAAGGACGUCGAUGAGUCCGAACAGGCACCAGCUUUGGAAAAGGUUA ((((.((((((...............(((....))).......(((((((...(........((((((((((((......))))).).)))))).)..)).)))))))))))...)))). ( -35.40) >DroSim_CAF1 113046 120 - 1 AGCUGUCCAAAAAGGAACGAAAAAAACUUAAGGAGGAAAAUAAGCUGGGCAAGGGCAAAAAGUCGGACCUCGACAAGGACGUCGAUGAGUCCGAACAGGCACCAGCUUUGGAAAAGGUUA ((((.((((((...............(((....))).......(((((((...(........((((((((((((......))))).).)))))).)..)).)))))))))))...)))). ( -35.40) >DroEre_CAF1 109409 120 - 1 AGUUGUCAAAAAAGGAGCGAAAAAAACUUAAGGAGGAAAAUAAGUUGGGCAAGGGCAAAAAGUCAGACCUCGAUAAGGACGUCGAUGAGUCUGAACAGGCACCAGCUUUGGAAAAGGUUA ..(((((.................((((((..........)))))).)))))((((......((((((((((((......))))).).))))))....)).))((((((....)))))). ( -27.17) >DroYak_CAF1 108196 120 - 1 AGUUGUCCAAAAAGGAGCGAAAAAAACUUAAGGAGGAAAAUAAGUUGGGCAAGGGCAAAAAGUCAGAUCUCGACAAGGACGUCGAUGAGUCUGAACAAGCACCAGCUUUGGAAAAAGUUA ..(((((((................(((((..........))))))))))))((((......((((((((((((......))))).)).)))))....)).))((((((....)))))). ( -29.09) >consensus AGCUGUCCAAAAAGGAACGAAAAAAACUUAAGGAGGAAAAUAAGCUGGGCAAGGGCAAAAAGUCGGACCUCGACAAGGACGUCGAUGAGUCCGAACAGGCACCAGCUUUGGAAAAGGUUA ((((.((((((...............(((....))).......(((((((...(........((((((((((((......))))).).)))))).)..)).)))))))))))...)))). (-30.38 = -29.02 + -1.36)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:24:59 2006