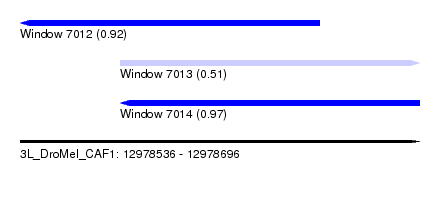
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,978,536 – 12,978,696 |
| Length | 160 |
| Max. P | 0.971555 |

| Location | 12,978,536 – 12,978,656 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.56 |
| Mean single sequence MFE | -30.90 |
| Consensus MFE | -22.95 |
| Energy contribution | -24.89 |
| Covariance contribution | 1.94 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916996 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12978536 120 - 23771897 AUACGGUGCGUAUGAGCGAUCCCUGCGAACAAUUAGUUGACGCAAUUUCUCAUAUGCUAACACCUCUCGUUGUCGCCGCAAUGCAAGCGAAGCCCCGGCGAUAUAAGUUGUAGCUCAAAA .(((((((((((((((.(((...(((((((.....)))..))))))).)))))))))....)))......((((((((....((.......))..))))))))......)))........ ( -32.60) >DroSim_CAF1 105547 120 - 1 AUACGGUGCGUAUGAGCGAUCCCUGUCAACAAUCAGUUGACGCAAUUUCUCAUAUGCUAACACCUCUCGUUGUCGCCGCAAAGCAAGCGAAACCCCGGCGAUAUAAGUUGUAGCUCAAAA .(((((((((((((((.(((...(((((((.....)))))))..))).)))))))))....)))......(((((((((.......))(......))))))))......)))........ ( -34.70) >DroEre_CAF1 101687 93 - 1 AUACGGUGCGUAUGAGCGAUCCCUGUCAACUAUCCGUUGACGCAAUUUCUCAUAUACCAACACCUCUCGU---------------------------ACGAUAUAAACUGUAGCUAAAAA .((((((.((((((((.(......((((((.....))))))(........)............).)))))---------------------------)))......))))))........ ( -20.80) >DroYak_CAF1 99956 120 - 1 AUACGGUGCGUAUGAGCGAUCCCUGUCAACUAUCAGUUGACGUAAUUUAUCAUAUGCCAACACCUCUCGUUGUUGCCGCAAAGCAACCAAGAGCCAGGCGAUAUAAACUGUAGCUCAAAA .((((((((((((((..(((..(.((((((.....)))))))..)))..))))))))...(.((((((...(((((......)))))...)))..))).)......))))))........ ( -35.50) >consensus AUACGGUGCGUAUGAGCGAUCCCUGUCAACAAUCAGUUGACGCAAUUUCUCAUAUGCCAACACCUCUCGUUGUCGCCGCAAAGCAAGCGAAACCCCGGCGAUAUAAACUGUAGCUCAAAA .(((((((((((((((.(((...(((((((.....)))))))..))).))))))))).............(((((((...................)))))))...))))))........ (-22.95 = -24.89 + 1.94)


| Location | 12,978,576 – 12,978,696 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.13 |
| Mean single sequence MFE | -37.82 |
| Consensus MFE | -29.72 |
| Energy contribution | -30.68 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.79 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.513182 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12978576 120 + 23771897 UGCGGCGACAACGAGAGGUGUUAGCAUAUGAGAAAUUGCGUCAACUAAUUGUUCGCAGGGAUCGCUCAUACGCACCGUAUUCCGGUACCAGUACCGGUUACGCUGCCGAACGUCAAAGUG ..((((((((.(....).)))).((.((((((...(((((.(((....)))..)))))......)))))).))..((((..((((((....)))))).))))..))))............ ( -39.30) >DroSec_CAF1 95933 103 + 1 -----------------GUGUUAGCAUAUGAGAAAUUGCGUCAACUGAUUGUUGACAGGGAUCGCUCAUACGCACCGUAUUCCGGUACCAGUACCGGUGACGCUGCCGAACGUCAAAGUG -----------------((((((((.((((((..(((.(((((((.....)))))).).)))..)))))).))........((((((....))))))))))))................. ( -35.70) >DroSim_CAF1 105587 120 + 1 UGCGGCGACAACGAGAGGUGUUAGCAUAUGAGAAAUUGCGUCAACUGAUUGUUGACAGGGAUCGCUCAUACGCACCGUAUUCCGGUACUAGUACCGGUGACGCUGCCGAACGUCAAAGUG (((((.((((.(....).)))).((.((((((..(((.(((((((.....)))))).).)))..)))))).)).)))))..((((((....))))))(((((........)))))..... ( -44.50) >DroEre_CAF1 101710 110 + 1 ----------ACGAGAGGUGUUGGUAUAUGAGAAAUUGCGUCAACGGAUAGUUGACAGGGAUCGCUCAUACGCACCGUAUUCCGGUACCUGUACCUGUAAAGCUGCCGAACGUCAAAGUG ----------....((.((.((((((((((((..(((.(((((((.....)))))).).)))..)))))).((....(((...((((....)))).)))..)))))))))).))...... ( -32.10) >DroYak_CAF1 99996 120 + 1 UGCGGCAACAACGAGAGGUGUUGGCAUAUGAUAAAUUACGUCAACUGAUAGUUGACAGGGAUCGCUCAUACGCACCGUAUUCCGGUAGCUGUACCUGCGACGCUGCCGAACGUCAAAGUG ((((((((((.(....).)))))((.(((((...((..(((((((.....)))))).)..))...))))).)).)))))...((((((((((....)))..)))))))............ ( -37.50) >consensus UGCGGC_ACAACGAGAGGUGUUAGCAUAUGAGAAAUUGCGUCAACUGAUUGUUGACAGGGAUCGCUCAUACGCACCGUAUUCCGGUACCAGUACCGGUGACGCUGCCGAACGUCAAAGUG ...........((...(((((((((.((((((..(((.(((((((.....)))))).).)))..)))))).))........((((((....)))))))))))))..))............ (-29.72 = -30.68 + 0.96)

| Location | 12,978,576 – 12,978,696 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.13 |
| Mean single sequence MFE | -38.64 |
| Consensus MFE | -29.40 |
| Energy contribution | -30.92 |
| Covariance contribution | 1.52 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.971555 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12978576 120 - 23771897 CACUUUGACGUUCGGCAGCGUAACCGGUACUGGUACCGGAAUACGGUGCGUAUGAGCGAUCCCUGCGAACAAUUAGUUGACGCAAUUUCUCAUAUGCUAACACCUCUCGUUGUCGCCGCA .....((.((..((((((((...((((((....)))))).....((((((((((((.(((...(((((((.....)))..))))))).)))))))))....)))...)))))))).)))) ( -40.50) >DroSec_CAF1 95933 103 - 1 CACUUUGACGUUCGGCAGCGUCACCGGUACUGGUACCGGAAUACGGUGCGUAUGAGCGAUCCCUGUCAACAAUCAGUUGACGCAAUUUCUCAUAUGCUAACAC----------------- .....((((((......))))))((((((....)))))).....(..(((((((((.(((...(((((((.....)))))))..))).)))))))))...)..----------------- ( -38.40) >DroSim_CAF1 105587 120 - 1 CACUUUGACGUUCGGCAGCGUCACCGGUACUAGUACCGGAAUACGGUGCGUAUGAGCGAUCCCUGUCAACAAUCAGUUGACGCAAUUUCUCAUAUGCUAACACCUCUCGUUGUCGCCGCA .....((.((..((((((((...((((((....)))))).....((((((((((((.(((...(((((((.....)))))))..))).)))))))))....)))...)))))))).)))) ( -44.50) >DroEre_CAF1 101710 110 - 1 CACUUUGACGUUCGGCAGCUUUACAGGUACAGGUACCGGAAUACGGUGCGUAUGAGCGAUCCCUGUCAACUAUCCGUUGACGCAAUUUCUCAUAUACCAACACCUCUCGU---------- ........................((((....((((((.....))))))(((((((.(((...(((((((.....)))))))..))).)))))))......)))).....---------- ( -28.80) >DroYak_CAF1 99996 120 - 1 CACUUUGACGUUCGGCAGCGUCGCAGGUACAGCUACCGGAAUACGGUGCGUAUGAGCGAUCCCUGUCAACUAUCAGUUGACGUAAUUUAUCAUAUGCCAACACCUCUCGUUGUUGCCGCA ............((((((((.((.((((......((((.....))))((((((((..(((..(.((((((.....)))))))..)))..))))))))....))))..)).)))))))).. ( -41.00) >consensus CACUUUGACGUUCGGCAGCGUCACCGGUACUGGUACCGGAAUACGGUGCGUAUGAGCGAUCCCUGUCAACAAUCAGUUGACGCAAUUUCUCAUAUGCUAACACCUCUCGUUGU_GCCGCA .....((((((......))))))((((((....)))))).....((((((((((((.(((...(((((((.....)))))))..))).)))))))))....)))................ (-29.40 = -30.92 + 1.52)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:24:49 2006