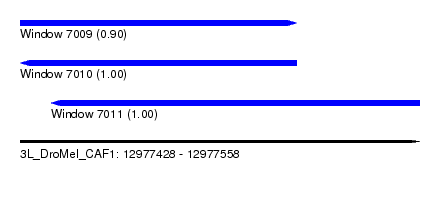
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,977,428 – 12,977,558 |
| Length | 130 |
| Max. P | 0.998971 |

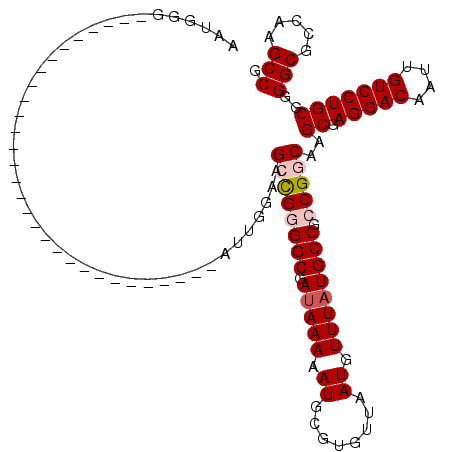
| Location | 12,977,428 – 12,977,518 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.23 |
| Mean single sequence MFE | -32.88 |
| Consensus MFE | -25.35 |
| Energy contribution | -26.16 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.904824 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12977428 90 + 23771897 AAUGGG------------------------------AUUGGGGGCCGGGGCGAUAAAAAUGCGUGUUAAUGUUUAUGCCGCCGGCAAGCGAGGACAAUUGUCCUGCGGGGCGUCAAGCCG ....((------------------------------.((((.(((((((((.((((....((((....))))))))))).)))))..((.(((((....)))))))....).)))).)). ( -34.50) >DroSec_CAF1 94825 90 + 1 UAUGGG------------------------------AUUGGAAGCCGGGGCGAUAAAAAUGCGUGUUAAUGUUUCUGCCGCCGGCAAGCGAGGACAAUUGUCCUGCGGGGCGCCAAGCCG ....((------------------------------.((((..((((((((((.......((((....)))).)).))).)))))..((.(((((....)))))))......)))).)). ( -34.00) >DroSim_CAF1 104460 90 + 1 UAUGGG------------------------------AUUGGAAGCCGGGGCGAUAAAAAUGCGUGUUAAUGUUUAUGCCGCCGGCAAGCGAGGACAAUUGUCCUGCGGGGCGCCAAGCCU ...(((------------------------------.((((..((((((((.((((....((((....))))))))))).)))))..((.(((((....)))))))......)))).))) ( -35.40) >DroEre_CAF1 100540 120 + 1 AAUGGGAAUGGGAAUGGGAAUGGGUAUGGGUAUGGGAAUGGAAGGUGUGGCGAUAAAAAUGCGUGUUAAUGUUUAUGCCGCCGGCGAGCGAGGACAAUUGUCCUGCGGGGCGCCAAGCCG .........((.....((....((((((..(((....(((.(...(((....)))....).)))....)))..)))))).))((((.((.(((((....)))))))....))))...)). ( -31.50) >DroYak_CAF1 98829 86 + 1 AAUGGGAAUGGG------------------------AAUGGAAGGUGUGGCGAUAAAAA----------UGUUUAUGCCGCCGGCAAGCGAGGACAAUUGUCCUGCGGGGCGCCAAGCCG ....((..(((.------------------------..........(((((.(((((..----------..))))))))))((.(..((.(((((....)))))))..).)))))..)). ( -29.00) >consensus AAUGGG______________________________AUUGGAAGCCGGGGCGAUAAAAAUGCGUGUUAAUGUUUAUGCCGCCGGCAAGCGAGGACAAUUGUCCUGCGGGGCGCCAAGCCG ...........................................((((((((.(((((.((........)).)))))))).)))))..((.(((((....)))))))..(((.....))). (-25.35 = -26.16 + 0.81)

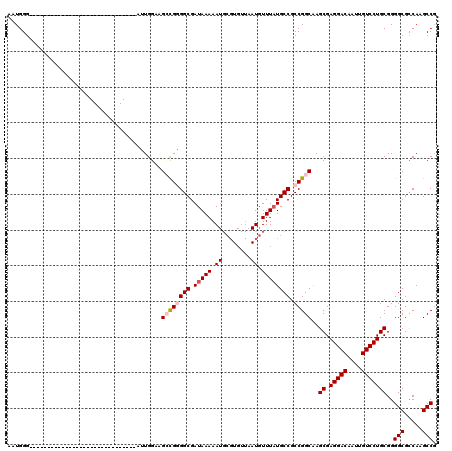
| Location | 12,977,428 – 12,977,518 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.23 |
| Mean single sequence MFE | -27.36 |
| Consensus MFE | -20.71 |
| Energy contribution | -22.16 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.76 |
| SVM decision value | 2.91 |
| SVM RNA-class probability | 0.997677 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12977428 90 - 23771897 CGGCUUGACGCCCCGCAGGACAAUUGUCCUCGCUUGCCGGCGGCAUAAACAUUAACACGCAUUUUUAUCGCCCCGGCCCCCAAU------------------------------CCCAUU .(((.....)))..(((((((....))))).))..(((((.((((((((.((........)).))))).)))))))).......------------------------------...... ( -28.80) >DroSec_CAF1 94825 90 - 1 CGGCUUGGCGCCCCGCAGGACAAUUGUCCUCGCUUGCCGGCGGCAGAAACAUUAACACGCAUUUUUAUCGCCCCGGCUUCCAAU------------------------------CCCAUA .((.((((......(((((((....))))).))..(((((.(((.((....................))))))))))..)))).------------------------------)).... ( -28.75) >DroSim_CAF1 104460 90 - 1 AGGCUUGGCGCCCCGCAGGACAAUUGUCCUCGCUUGCCGGCGGCAUAAACAUUAACACGCAUUUUUAUCGCCCCGGCUUCCAAU------------------------------CCCAUA .((.((((......(((((((....))))).))..(((((.((((((((.((........)).))))).))))))))..)))).------------------------------)).... ( -31.00) >DroEre_CAF1 100540 120 - 1 CGGCUUGGCGCCCCGCAGGACAAUUGUCCUCGCUCGCCGGCGGCAUAAACAUUAACACGCAUUUUUAUCGCCACACCUUCCAUUCCCAUACCCAUACCCAUUCCCAUUCCCAUUCCCAUU .(((.(((((...((.(((((....)))))))..)))))(((...............))).........)))................................................ ( -24.46) >DroYak_CAF1 98829 86 - 1 CGGCUUGGCGCCCCGCAGGACAAUUGUCCUCGCUUGCCGGCGGCAUAAACA----------UUUUUAUCGCCACACCUUCCAUU------------------------CCCAUUCCCAUU .((..(((.(((..(((((((....))))).)).....)))((((((((..----------..))))).)))............------------------------.)))..)).... ( -23.80) >consensus CGGCUUGGCGCCCCGCAGGACAAUUGUCCUCGCUUGCCGGCGGCAUAAACAUUAACACGCAUUUUUAUCGCCCCGGCUUCCAAU______________________________CCCAUU .(((.....)))..(((((((....))))).))..(((((.((((((((.((........)).))))).))))))))........................................... (-20.71 = -22.16 + 1.45)

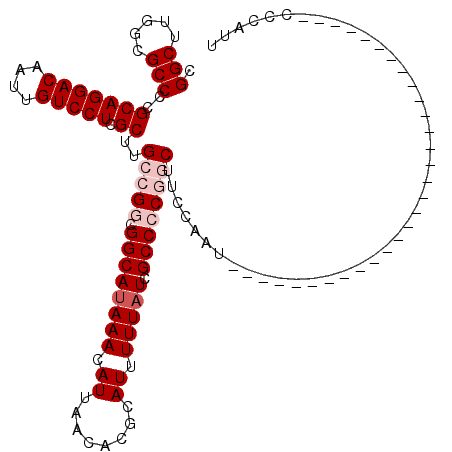
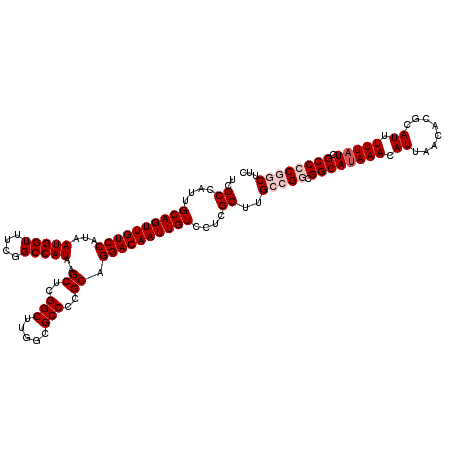
| Location | 12,977,438 – 12,977,558 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.17 |
| Mean single sequence MFE | -39.67 |
| Consensus MFE | -35.29 |
| Energy contribution | -36.74 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.89 |
| SVM decision value | 3.31 |
| SVM RNA-class probability | 0.998971 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12977438 120 - 23771897 UCGCCAUUGCAGUUGUCCAUAAUGGUUUCGGCCAUAAGCUCGGCUUGACGCCCCGCAGGACAAUUGUCCUCGCUUGCCGGCGGCAUAAACAUUAACACGCAUUUUUAUCGCCCCGGCCCC ..((....((((((((((...(((((....)))))..((..(((.....)))..)).))))))))))....))..(((((.((((((((.((........)).))))).))))))))... ( -43.30) >DroSec_CAF1 94835 120 - 1 UCGCCAUUGCAGUUGUCCAUAAUGGUUUCGGCCAUAAGCUCGGCUUGGCGCCCCGCAGGACAAUUGUCCUCGCUUGCCGGCGGCAGAAACAUUAACACGCAUUUUUAUCGCCCCGGCUUC ..((....((((((((((...(((((....)))))..((..(((.....)))..)).))))))))))....))..(((((.(((.((....................))))))))))... ( -41.35) >DroSim_CAF1 104470 120 - 1 UCGCCAUUGCAGUUGUCCAUAAUGGUUUCGGCCAUAAGCUAGGCUUGGCGCCCCGCAGGACAAUUGUCCUCGCUUGCCGGCGGCAUAAACAUUAACACGCAUUUUUAUCGCCCCGGCUUC ..((....((((((((((...(((((....)))))..((..(((.....)))..)).))))))))))....))..(((((.((((((((.((........)).))))).))))))))... ( -44.00) >DroEre_CAF1 100580 120 - 1 UCGCCAUUGCAGUUGUCCAUAAUGGUUUCGGCCAUAAGCUCGGCUUGGCGCCCCGCAGGACAAUUGUCCUCGCUCGCCGGCGGCAUAAACAUUAACACGCAUUUUUAUCGCCACACCUUC ..(((...((((((((((...(((((....)))))..((..(((.....)))..)).))))))))))....(....).)))((((((((.((........)).))))).)))........ ( -34.50) >DroYak_CAF1 98845 110 - 1 UCGCCAUUGCAGUUGUCCAUAAUGGUUUCGGCCAUAAGCUCGGCUUGGCGCCCCGCAGGACAAUUGUCCUCGCUUGCCGGCGGCAUAAACA----------UUUUUAUCGCCACACCUUC ..(((...((((((((((...(((((....)))))..((..(((.....)))..)).))))))))))....(....).)))((((((((..----------..))))).)))........ ( -35.20) >consensus UCGCCAUUGCAGUUGUCCAUAAUGGUUUCGGCCAUAAGCUCGGCUUGGCGCCCCGCAGGACAAUUGUCCUCGCUUGCCGGCGGCAUAAACAUUAACACGCAUUUUUAUCGCCCCGGCUUC ..((....((((((((((...(((((....)))))..((..(((.....)))..)).))))))))))....))..(((((.((((((((.((........)).))))).))))))))... (-35.29 = -36.74 + 1.45)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:24:46 2006