
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,976,714 – 12,976,858 |
| Length | 144 |
| Max. P | 0.999254 |

| Location | 12,976,714 – 12,976,818 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.75 |
| Mean single sequence MFE | -29.88 |
| Consensus MFE | -28.56 |
| Energy contribution | -28.44 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.96 |
| SVM decision value | 3.38 |
| SVM RNA-class probability | 0.999121 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12976714 104 + 23771897 ----------------GUGUUUGAUGGCAUUCGAUUGGCAUUUUCAAUGUAUAUAAGUGGCUGCGUGUGGGGUGAAAAGUUAUUGCAUCAUAAUUUCCCCCACUAAGCAUUUAUAUUGCG ----------------.((...((((.((......)).))))..))..(((((((((((.((....((((((.(((..(((((......))))))))))))))..)))))))))).))). ( -29.00) >DroSec_CAF1 94029 104 + 1 ----------------GUGUUUGAUGGCAUUCGAUUGGCAUUUUCAAUGUAUGUAAGUGGCUGCGUGUGGGGUGAAAAGUUAUUGCAUCAUAAUUUCCCCCACUAAGCAUUUAUAUUGCA ----------------.((...((((.((......)).))))..)).((((((((((((.((....((((((.(((..(((((......))))))))))))))..)))))))))).)))) ( -29.20) >DroSim_CAF1 103728 104 + 1 ----------------GUGUUUGAUGGCAUUCGAUUGGCAUUUUCAAUGUAUGUAAGUGGCUGCGUGUGGGGUGAAAAGUUAUUGCAUCAUAAUUUCCCCCACUAAGCAUUUAUAUUGCA ----------------.((...((((.((......)).))))..)).((((((((((((.((....((((((.(((..(((((......))))))))))))))..)))))))))).)))) ( -29.20) >DroEre_CAF1 99804 120 + 1 GCUUGUGUUUGUGCUUGUGUUUGAUGGCAUUCGAUUGGCAUUUUCAAUGUAUGUAAGUGGCUGCGUGUGGGGUGAAAAGUUAUUGCAUCAUAAUUUCCCCCACUAAGCAUUUAUAUUGCA ((..((((..(((((((((...((((.((......)).))))..))(((((.((.....)))))))((((((.(((..(((((......)))))))))))))))))))))..)))).)). ( -33.70) >DroYak_CAF1 98033 102 + 1 ------------------GUUUGAUGGCAUUCGAUUGGCAUUUUCAAUGUAUGUAAGUGGCUGCGUGUGGGGUGAAAAGUUAUUGCAUCAUAAUUUCCCCCACUAAGCAUUUAUAUUGCA ------------------(...((((.((......)).))))..)..((((((((((((.((....((((((.(((..(((((......))))))))))))))..)))))))))).)))) ( -28.30) >consensus ________________GUGUUUGAUGGCAUUCGAUUGGCAUUUUCAAUGUAUGUAAGUGGCUGCGUGUGGGGUGAAAAGUUAUUGCAUCAUAAUUUCCCCCACUAAGCAUUUAUAUUGCA .................((...((((.((......)).))))..)).((((((((((((.((....((((((.(((..(((((......))))))))))))))..)))))))))).)))) (-28.56 = -28.44 + -0.12)

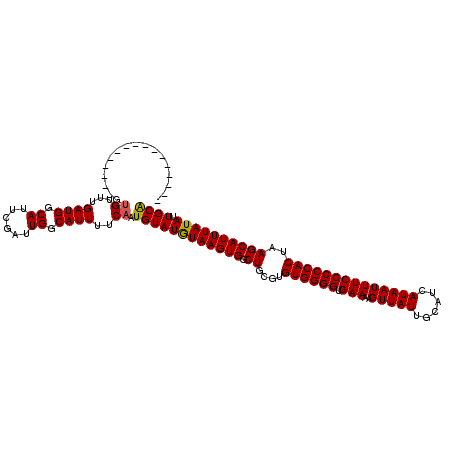
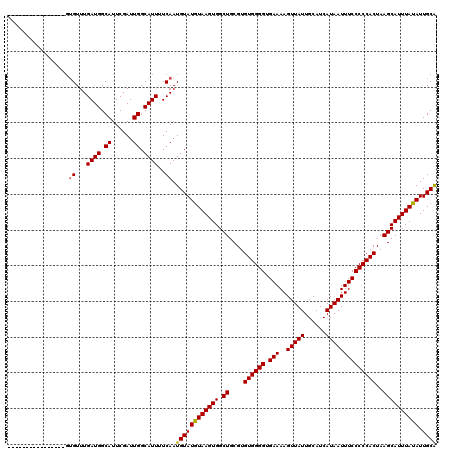
| Location | 12,976,714 – 12,976,818 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.75 |
| Mean single sequence MFE | -19.80 |
| Consensus MFE | -19.16 |
| Energy contribution | -19.16 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.988594 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12976714 104 - 23771897 CGCAAUAUAAAUGCUUAGUGGGGGAAAUUAUGAUGCAAUAACUUUUCACCCCACACGCAGCCACUUAUAUACAUUGAAAAUGCCAAUCGAAUGCCAUCAAACAC---------------- .(((((((((.(((...((((((((((((((......))))..)))).))))))..))).....))))))..((((.......))))....)))..........---------------- ( -20.70) >DroSec_CAF1 94029 104 - 1 UGCAAUAUAAAUGCUUAGUGGGGGAAAUUAUGAUGCAAUAACUUUUCACCCCACACGCAGCCACUUACAUACAUUGAAAAUGCCAAUCGAAUGCCAUCAAACAC---------------- .(((.......(((...((((((((((((((......))))..)))).))))))..))).............((((.......))))....)))..........---------------- ( -19.10) >DroSim_CAF1 103728 104 - 1 UGCAAUAUAAAUGCUUAGUGGGGGAAAUUAUGAUGCAAUAACUUUUCACCCCACACGCAGCCACUUACAUACAUUGAAAAUGCCAAUCGAAUGCCAUCAAACAC---------------- .(((.......(((...((((((((((((((......))))..)))).))))))..))).............((((.......))))....)))..........---------------- ( -19.10) >DroEre_CAF1 99804 120 - 1 UGCAAUAUAAAUGCUUAGUGGGGGAAAUUAUGAUGCAAUAACUUUUCACCCCACACGCAGCCACUUACAUACAUUGAAAAUGCCAAUCGAAUGCCAUCAAACACAAGCACAAACACAAGC ...........(((((.((((((((((((((......))))..)))).)))))).............(((.(((((.......)))).).)))...........)))))........... ( -21.00) >DroYak_CAF1 98033 102 - 1 UGCAAUAUAAAUGCUUAGUGGGGGAAAUUAUGAUGCAAUAACUUUUCACCCCACACGCAGCCACUUACAUACAUUGAAAAUGCCAAUCGAAUGCCAUCAAAC------------------ .(((.......(((...((((((((((((((......))))..)))).))))))..))).............((((.......))))....)))........------------------ ( -19.10) >consensus UGCAAUAUAAAUGCUUAGUGGGGGAAAUUAUGAUGCAAUAACUUUUCACCCCACACGCAGCCACUUACAUACAUUGAAAAUGCCAAUCGAAUGCCAUCAAACAC________________ .(((.......(((...((((((((((((((......))))..)))).))))))..))).............((((.......))))....))).......................... (-19.16 = -19.16 + 0.00)

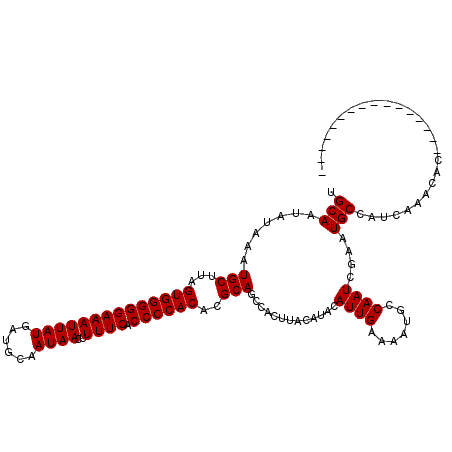
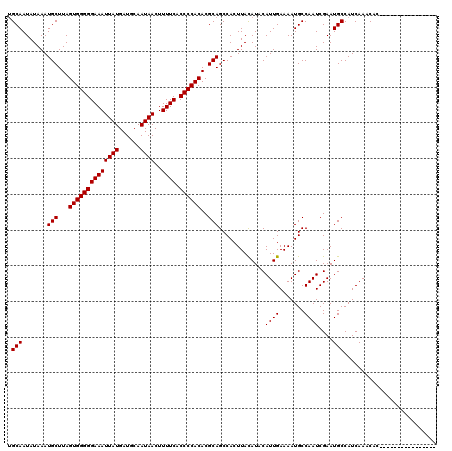
| Location | 12,976,738 – 12,976,858 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.17 |
| Mean single sequence MFE | -29.83 |
| Consensus MFE | -29.85 |
| Energy contribution | -29.53 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.07 |
| Structure conservation index | 1.00 |
| SVM decision value | 3.22 |
| SVM RNA-class probability | 0.998773 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12976738 120 + 23771897 UUUUCAAUGUAUAUAAGUGGCUGCGUGUGGGGUGAAAAGUUAUUGCAUCAUAAUUUCCCCCACUAAGCAUUUAUAUUGCGCAUAAUUACAUCCGCACAUGCCCACGAUGCCGUCUAAAAA ........((((....((((..((((((((((((...((((((((((..((((.................))))..)))).)))))).))))).))))))))))).)))).......... ( -30.73) >DroSec_CAF1 94053 120 + 1 UUUUCAAUGUAUGUAAGUGGCUGCGUGUGGGGUGAAAAGUUAUUGCAUCAUAAUUUCCCCCACUAAGCAUUUAUAUUGCACAUAAUUACAUCCGCACAUGCCCACGAUGCCGUCUAUAAA ...........((((..((((.((((((((((((...((((((((((..((((.................))))..)))).)))))).))))).))))))).......))))..)))).. ( -30.23) >DroSim_CAF1 103752 120 + 1 UUUUCAAUGUAUGUAAGUGGCUGCGUGUGGGGUGAAAAGUUAUUGCAUCAUAAUUUCCCCCACUAAGCAUUUAUAUUGCACAUAAUUACAUCCGCACAUGCCCACGAUGCCGUCUAUAAA ...........((((..((((.((((((((((((...((((((((((..((((.................))))..)))).)))))).))))).))))))).......))))..)))).. ( -30.23) >DroEre_CAF1 99844 120 + 1 UUUUCAAUGUAUGUAAGUGGCUGCGUGUGGGGUGAAAAGUUAUUGCAUCAUAAUUUCCCCCACUAAGCAUUUAUAUUGCACAUAAUUACAUCCACACAUGCCCACGAUGCCGUCUAUAAA ...........((((..((((.((((((((((((...((((((((((..((((.................))))..)))).)))))).))))).))))))).......))))..)))).. ( -30.23) >DroYak_CAF1 98055 120 + 1 UUUUCAAUGUAUGUAAGUGGCUGCGUGUGGGGUGAAAAGUUAUUGCAUCAUAAUUUCCCCCACUAAGCAUUUAUAUUGCACAUAAUUACAUUCACACAUGCCCACGAUGCCGUCUAUAAA ...........((((..((((....(((((((((((.((((((((((..((((.................))))..)))).))))))...))))).....))))))..))))..)))).. ( -27.73) >consensus UUUUCAAUGUAUGUAAGUGGCUGCGUGUGGGGUGAAAAGUUAUUGCAUCAUAAUUUCCCCCACUAAGCAUUUAUAUUGCACAUAAUUACAUCCGCACAUGCCCACGAUGCCGUCUAUAAA ........((((....((((..((((((((((((...((((((((((..((((.................))))..)))).)))))).))))).))))))))))).)))).......... (-29.85 = -29.53 + -0.32)

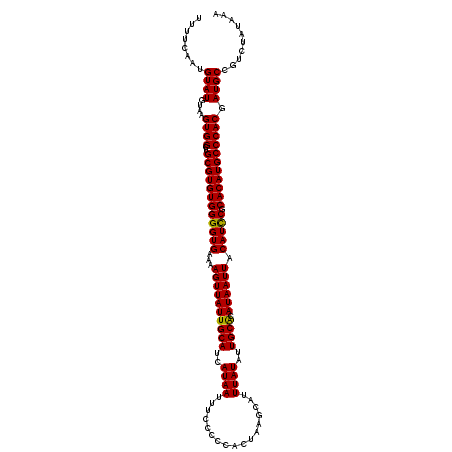

| Location | 12,976,738 – 12,976,858 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.17 |
| Mean single sequence MFE | -32.82 |
| Consensus MFE | -32.56 |
| Energy contribution | -32.16 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.99 |
| SVM decision value | 3.46 |
| SVM RNA-class probability | 0.999254 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12976738 120 - 23771897 UUUUUAGACGGCAUCGUGGGCAUGUGCGGAUGUAAUUAUGCGCAAUAUAAAUGCUUAGUGGGGGAAAUUAUGAUGCAAUAACUUUUCACCCCACACGCAGCCACUUAUAUACAUUGAAAA .........(((..(((((((((.((((.(((....))).))))......)))))).((((((((((((((......))))..)))).)))))))))..))).................. ( -34.90) >DroSec_CAF1 94053 120 - 1 UUUAUAGACGGCAUCGUGGGCAUGUGCGGAUGUAAUUAUGUGCAAUAUAAAUGCUUAGUGGGGGAAAUUAUGAUGCAAUAACUUUUCACCCCACACGCAGCCACUUACAUACAUUGAAAA .........(((..(((((((((.((((.(((....))).))))......)))))).((((((((((((((......))))..)))).)))))))))..))).................. ( -33.30) >DroSim_CAF1 103752 120 - 1 UUUAUAGACGGCAUCGUGGGCAUGUGCGGAUGUAAUUAUGUGCAAUAUAAAUGCUUAGUGGGGGAAAUUAUGAUGCAAUAACUUUUCACCCCACACGCAGCCACUUACAUACAUUGAAAA .........(((..(((((((((.((((.(((....))).))))......)))))).((((((((((((((......))))..)))).)))))))))..))).................. ( -33.30) >DroEre_CAF1 99844 120 - 1 UUUAUAGACGGCAUCGUGGGCAUGUGUGGAUGUAAUUAUGUGCAAUAUAAAUGCUUAGUGGGGGAAAUUAUGAUGCAAUAACUUUUCACCCCACACGCAGCCACUUACAUACAUUGAAAA .........(((..(((((((((.((((..((((......)))).)))).)))))).((((((((((((((......))))..)))).)))))))))..))).................. ( -32.00) >DroYak_CAF1 98055 120 - 1 UUUAUAGACGGCAUCGUGGGCAUGUGUGAAUGUAAUUAUGUGCAAUAUAAAUGCUUAGUGGGGGAAAUUAUGAUGCAAUAACUUUUCACCCCACACGCAGCCACUUACAUACAUUGAAAA .........(((..(((((((((.((((..((((......)))).)))).)))))).((((((((((((((......))))..)))).)))))))))..))).................. ( -30.60) >consensus UUUAUAGACGGCAUCGUGGGCAUGUGCGGAUGUAAUUAUGUGCAAUAUAAAUGCUUAGUGGGGGAAAUUAUGAUGCAAUAACUUUUCACCCCACACGCAGCCACUUACAUACAUUGAAAA .........(((..(((((((((.((((.(((....))).))))......)))))).((((((((((((((......))))..)))).)))))))))..))).................. (-32.56 = -32.16 + -0.40)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:24:43 2006