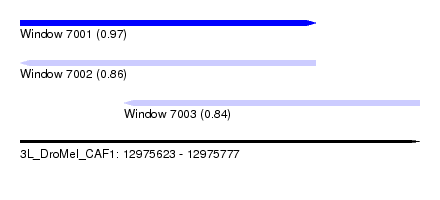
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,975,623 – 12,975,777 |
| Length | 154 |
| Max. P | 0.966107 |

| Location | 12,975,623 – 12,975,737 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.63 |
| Mean single sequence MFE | -33.59 |
| Consensus MFE | -22.44 |
| Energy contribution | -25.52 |
| Covariance contribution | 3.08 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.966107 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
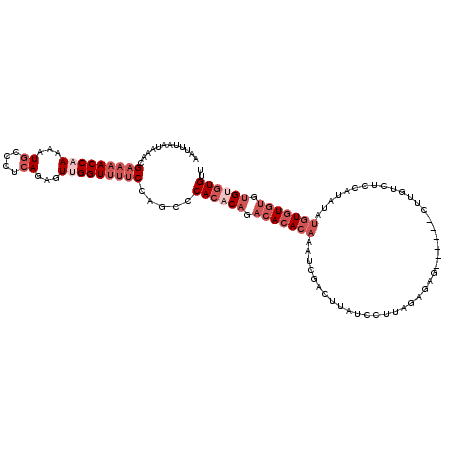
>3L_DroMel_CAF1 12975623 114 + 23771897 UUUUUAGAUCCCAGUCCAAGCAAUUGAAUGAUGUCAGUCAAACACACACACACACACAUAUGGAGACAAG------GUCUCUAAGGAUAAGUCGAUUUGUGUGUCUGUGUGGGCUGGAAA ..........(((((((......((((.((....)).))))....(((((.(((((((..(((((((...------)))))))..((....))....))))))).))))))))))))... ( -38.40) >DroSec_CAF1 92996 114 + 1 UUUUUAGAUCCCAGUCCAAGCAAUUGAAUGAUGUCAGUCAAACACACACACACACAUAUAUGGAGACAAG------CUCUCUAAGGAUAAGUCGAUUUGUGUGUCUGUGUGGGCUGGAAA ..........(((((((......((((.((....)).))))..(((((.(((((((....((((((....------.))))))..((....))....))))))).))))))))))))... ( -36.50) >DroSim_CAF1 102703 110 + 1 UUUUUAGAUCCCAGUCCAAGCAAUUGAAUGAUGUCAGUCAAACACUC----ACACAUAUAUGGAGACAAG------CUCUCUAAGGAUAAGUCGAUUUGUGUGUCUGUGUGGGCUGGAAA ..........(((((((......((((.((....)).))))((((..----(((((((..((((((....------.))))))..((....))....)))))))..)))))))))))... ( -33.00) >DroEre_CAF1 98736 114 + 1 UUUUUAGAUCCAGGUCCAAGGAAUUGAAUGGUGCCAGUCAAACACCCACACACACACA----GAAGCAAGCGUUCUCUUUCUAAGGAUAAGUCGAUUUGUGUGUAUGUGUGGGCUAG--G .........((.((((((.((..((((.((....)).))))...))((((.(((((((----((.......(((((.......))))).......))))))))).)))))))))).)--) ( -30.84) >DroYak_CAF1 96932 101 + 1 UUUUUAGACCCAAGUCCAAGCAAUUGAAUGGUGCCAGUCAAACAGACACACACA-------------------UCUCUUUCUAAGGAUAAGUCGAUUUGUCUGUCUGUGUGGGCUGGAAA .......(((...((.(((....))).))))).((((((..(((((((.(((.(-------------------((.(((((....)).)))..))).))).)))))))...))))))... ( -29.20) >consensus UUUUUAGAUCCCAGUCCAAGCAAUUGAAUGAUGUCAGUCAAACACACACACACACACAUAUGGAGACAAG______CUCUCUAAGGAUAAGUCGAUUUGUGUGUCUGUGUGGGCUGGAAA ..........(((((((......((((.((....)).))))(((((.(((((((......((((((...........))))))..((....))....))))))).))))))))))))... (-22.44 = -25.52 + 3.08)

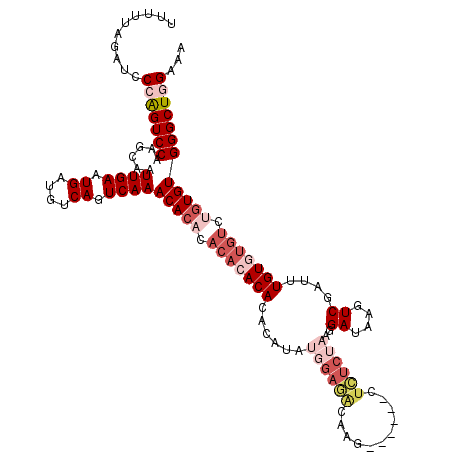

| Location | 12,975,623 – 12,975,737 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.63 |
| Mean single sequence MFE | -31.08 |
| Consensus MFE | -17.18 |
| Energy contribution | -19.86 |
| Covariance contribution | 2.68 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.858318 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12975623 114 - 23771897 UUUCCAGCCCACACAGACACACAAAUCGACUUAUCCUUAGAGAC------CUUGUCUCCAUAUGUGUGUGUGUGUGUGUUUGACUGACAUCAUUCAAUUGCUUGGACUGGGAUCUAAAAA .((((((((((((((.(((((((....((....))....(((((------...)))))....))))))).))))))...((((.((....)).))))......)).))))))........ ( -35.60) >DroSec_CAF1 92996 114 - 1 UUUCCAGCCCACACAGACACACAAAUCGACUUAUCCUUAGAGAG------CUUGUCUCCAUAUAUGUGUGUGUGUGUGUUUGACUGACAUCAUUCAAUUGCUUGGACUGGGAUCUAAAAA .((((((((((((((.(((((((....((....))....((((.------....))))......))))))).)))))).((((.((....)).))))......)).))))))........ ( -33.70) >DroSim_CAF1 102703 110 - 1 UUUCCAGCCCACACAGACACACAAAUCGACUUAUCCUUAGAGAG------CUUGUCUCCAUAUAUGUGU----GAGUGUUUGACUGACAUCAUUCAAUUGCUUGGACUGGGAUCUAAAAA .((((((.(((......((((((....((....))....((((.------....))))......)))))----)(((..((((.((....)).))))..)))))).))))))........ ( -25.00) >DroEre_CAF1 98736 114 - 1 C--CUAGCCCACACAUACACACAAAUCGACUUAUCCUUAGAAAGAGAACGCUUGCUUC----UGUGUGUGUGUGGGUGUUUGACUGGCACCAUUCAAUUCCUUGGACCUGGAUCUAAAAA .--...((((((((((((((.......((....))...((((.(((....)))..)))----)))))))))))))))..((((.((....)).)))).(((........)))........ ( -32.90) >DroYak_CAF1 96932 101 - 1 UUUCCAGCCCACACAGACAGACAAAUCGACUUAUCCUUAGAAAGAGA-------------------UGUGUGUGUCUGUUUGACUGGCACCAUUCAAUUGCUUGGACUUGGGUCUAAAAA ...((((..((.(((((((.(((.(((..(((.((....))))).))-------------------).))).))))))).)).))))..............((((((....))))))... ( -28.20) >consensus UUUCCAGCCCACACAGACACACAAAUCGACUUAUCCUUAGAGAG______CUUGUCUCCAUAUAUGUGUGUGUGUGUGUUUGACUGACAUCAUUCAAUUGCUUGGACUGGGAUCUAAAAA .((((((((((((((.(((((((.........................................))))))).)))))).((((.((....)).))))......)).))))))........ (-17.18 = -19.86 + 2.68)

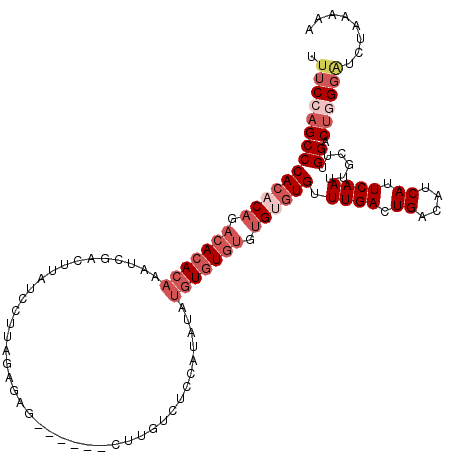
| Location | 12,975,663 – 12,975,777 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.15 |
| Mean single sequence MFE | -28.22 |
| Consensus MFE | -15.54 |
| Energy contribution | -18.54 |
| Covariance contribution | 3.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.836214 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12975663 114 - 23771897 AAUUUAAUAAACGAAAACCAAAAAUGCCCUCAGAGUAGGUUUUCCAGCCCACACAGACACACAAAUCGACUUAUCCUUAGAGAC------CUUGUCUCCAUAUGUGUGUGUGUGUGUGUU ............(((((((.....((....)).....))))))).(((.((((((.(((((((....((....))....(((((------...)))))....))))))).)))))).))) ( -31.40) >DroSec_CAF1 93036 114 - 1 AAAUUAAUAAACGAAAACCAAAAAUGCCCUAAGGGUUGGUUUUCCAGCCCACACAGACACACAAAUCGACUUAUCCUUAGAGAG------CUUGUCUCCAUAUAUGUGUGUGUGUGUGUU ............((((((((.....((((...)))))))))))).....((((((.(((((((....((....))....((((.------....))))......))))))).)))))).. ( -33.60) >DroSim_CAF1 102743 110 - 1 AAUUUAAUAAACGAAAACCAAAAAUGCCCUCAGAGUUGGUUUUCCAGCCCACACAGACACACAAAUCGACUUAUCCUUAGAGAG------CUUGUCUCCAUAUAUGUGU----GAGUGUU ............(((((((((...((....))...)))))))))......((((...((((((....((....))....((((.------....))))......)))))----).)))). ( -21.40) >DroEre_CAF1 98776 114 - 1 AAUUUAAUAACCGAAAACCAAAAAUGCCCUCAGAGUAGGUC--CUAGCCCACACAUACACACAAAUCGACUUAUCCUUAGAAAGAGAACGCUUGCUUC----UGUGUGUGUGUGGGUGUU ...........................(((......)))..--...((((((((((((((.......((....))...((((.(((....)))..)))----)))))))))))))))... ( -28.00) >DroYak_CAF1 96972 101 - 1 AAUUUAAUAACCGAAAACCAAAAAUGCCCUCAGAGUUGGUUUUCCAGCCCACACAGACAGACAAAUCGACUUAUCCUUAGAAAGAGA-------------------UGUGUGUGUCUGUU ............(((((((((...((....))...)))))))))........(((((((.(((.(((..(((.((....))))).))-------------------).))).))))))). ( -26.70) >consensus AAUUUAAUAAACGAAAACCAAAAAUGCCCUCAGAGUUGGUUUUCCAGCCCACACAGACACACAAAUCGACUUAUCCUUAGAGAG______CUUGUCUCCAUAUAUGUGUGUGUGUGUGUU ............(((((((((...((....))...))))))))).....((((((.(((((((.........................................))))))).)))))).. (-15.54 = -18.54 + 3.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:24:38 2006