| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,972,932 – 12,973,120 |
| Length | 188 |
| Max. P | 0.879449 |

| Location | 12,972,932 – 12,973,045 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.56 |
| Mean single sequence MFE | -31.44 |
| Consensus MFE | -25.00 |
| Energy contribution | -25.08 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.879449 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
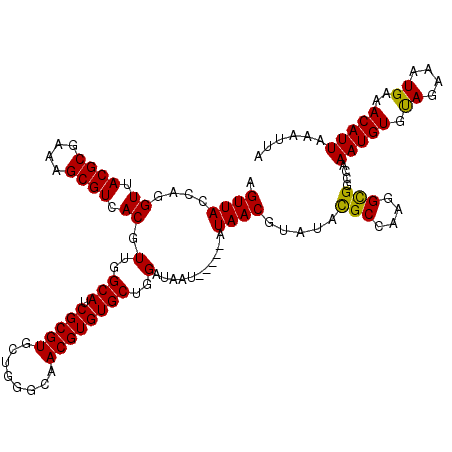
>3L_DroMel_CAF1 12972932 113 + 23771897 UA----UAACGUAAUAUAUAUGGGCAGGU---GUACUUGCAUUAUAUUUUUGCCAACUCUGUUGAGGCUAAAAGAACACUAGUUACCAGGUUACGCGAAAGCGUCACGUUGGCAUCGCGU ..----..((((...(((((...(((((.---...)))))..)))))...(((((((.(((..(..((((.........))))..))))((.((((....)))).)))))))))..)))) ( -34.20) >DroSec_CAF1 90315 113 + 1 UA----UAACGUAAUAUAUAUGGGCAGGU---GUACUUGCAUUAUAUUUAUGCCAACUCUAUUGAGGCUAAAAGAACACUAGUUACCAGGUUACGCGAAAGCGUCACGUUGGCAUCGCGU ..----..((((...(((((...(((((.---...)))))..)))))..((((((((........(((((.........))))).....((.((((....)))).)))))))))).)))) ( -33.30) >DroSim_CAF1 100009 113 + 1 UA----UAACGUAAUAUAUAUGGGCAAGU---GUACUUGCAUUAUAUUUAUGCCAACUCUAUUGAGGCUAAAAGAACACUAGUUACCAGGUUACGCGAAAGCGUCACGUUGGCAUCGCGU ..----..((((...(((((...(((((.---...)))))..)))))..((((((((........(((((.........))))).....((.((((....)))).)))))))))).)))) ( -33.60) >DroEre_CAF1 95690 119 + 1 UAGUAAUUUCUCUAUAUAUGUGUGUAAUUUCCUUACUUGCAUGAUCACCUUGCCAACUCUAUUGAGGCCAGAAGAACUCGCGUUACCAGGUUACGCGA-AGCGUCACGUUGGCAUCGCGU ..((............((((((.((((.....)))).)))))).......((((((((((.(((....))).)))..((((((.((...)).))))))-........)))))))..)).. ( -26.20) >DroYak_CAF1 94264 101 + 1 UA----CAUCGCUAUAUAUGUGA---------------GCAUGAUCACUUUGCCAACUCUAUUGAGGCUAGAAGAAUACUCGUUACCAGGUUACGCGAAAGCGUCACGUUGGCAUCGCGU ..----...(((.......((((---------------......))))..(((((((......(((............)))........((.((((....)))).)))))))))..))). ( -29.90) >consensus UA____UAACGUAAUAUAUAUGGGCAAGU___GUACUUGCAUUAUAUUUUUGCCAACUCUAUUGAGGCUAAAAGAACACUAGUUACCAGGUUACGCGAAAGCGUCACGUUGGCAUCGCGU .........(((...........((((((.....))))))..........((((((((((.(((....))).)))..............((.((((....)))).)))))))))..))). (-25.00 = -25.08 + 0.08)


| Location | 12,973,005 – 12,973,120 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.84 |
| Mean single sequence MFE | -32.73 |
| Consensus MFE | -29.44 |
| Energy contribution | -29.12 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.653571 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12973005 115 + 23771897 AGUUACCAGGUUACGCGAAAGCGUCACGUUGGCAUCGCGUGCUGGGCAACGUGUGCUGGAUAAU-----AUAACGUAUACGCCAAGGCGGCCAAAUGUGUAGAAAUGAAACAUUAAAUUA .(((.((((((.((((....)))).)).))))...(((((..(((.(...((((((........-----.....))))))((....))).))).))))).........)))......... ( -31.82) >DroSec_CAF1 90388 115 + 1 AGUUACCAGGUUACGCGAAAGCGUCACGUUGGCAUCGCGUGCUGGGCAACGUGUGCUGGAUAAU-----AUAACGUAUAUGCCAAGGCGGCCAAAUGUGUAGAAAUGAAACAUUAAAUUA .((..((..((.((((....)))).)).(((((((.((((....(....)((((........))-----)).))))..)))))))))..))..(((((.((....))..)))))...... ( -30.70) >DroSim_CAF1 100082 115 + 1 AGUUACCAGGUUACGCGAAAGCGUCACGUUGGCAUCGCGUGCUGGGCAACGUGUGCUGGAUAAU-----AUAACGUAUACGCCAAGGCGGCCAAAUGUGUAGAAAUGAAACAUUAAAUUA .(((.((((((.((((....)))).)).))))...(((((..(((.(...((((((........-----.....))))))((....))).))).))))).........)))......... ( -31.82) >DroEre_CAF1 95770 113 + 1 CGUUACCAGGUUACGCGA-AGCGUCACGUUGGCAUCGCGUGCUU-ACAACGUGUGCUGGGUAGU-----AUAACGUAUACGCCAAGGUGGCCAAAUGUGUAGAAAUGAAACAUUAAAUUA .(((..((..(((((((.-...(((((.(((((...(((..(..-.....)..)))......((-----(((...)))))))))).)))))....)))))))...)).)))......... ( -32.40) >DroYak_CAF1 94325 119 + 1 CGUUACCAGGUUACGCGAAAGCGUCACGUUGGCAUCGCGUGCUU-ACAACGUGUGCUGGGUAGUGCGAUAUAACGUAUACGCCAAGGCGGCCAAAUGUGCAGAAAUGAAACAUUAAAUUA ...((((..((.((((....)))).))...((((.(((((....-...))))))))).))))(((((......))))).(((....)))....(((((.((....))..)))))...... ( -36.90) >consensus AGUUACCAGGUUACGCGAAAGCGUCACGUUGGCAUCGCGUGCUGGGCAACGUGUGCUGGAUAAU_____AUAACGUAUACGCCAAGGCGGCCAAAUGUGUAGAAAUGAAACAUUAAAUUA .((((....((.((((....)))).)).(..(((.(((((........))))))))..)...........)))).....(((....)))....(((((...........)))))...... (-29.44 = -29.12 + -0.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:24:29 2006