
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,970,139 – 12,970,379 |
| Length | 240 |
| Max. P | 0.995883 |

| Location | 12,970,139 – 12,970,259 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.25 |
| Mean single sequence MFE | -44.52 |
| Consensus MFE | -35.14 |
| Energy contribution | -36.34 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.79 |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.985591 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12970139 120 + 23771897 GUUUGUGCGAAGCCAUGGCCGUCCAGACGCCACUUGUCCUGAUAGUGGUGGUGAUGUACUCAAAUAUCGGUGGCACCCUCACGCCAGUGGGUGAUCCACCCAAUGUGAUCAUUGCCACCA ....(((((..(((((.((..((..((((.....))))..))..)).)))))..))))).........(((((((...(((((....((((((...)))))).)))))....))))))). ( -47.20) >DroSec_CAF1 87483 120 + 1 GUUUGUGUGAAGCCAUGGCCGUGCAGACGCCACUUGUCCUGAUAGUGGUGGUGAUGUACUCAAAUAUCGGUGGAACCCUCACUCCAGUGGGAGAUCCACCCAAUGUGAUCAUUGCCACCA (((((..((..((....))))..))))).................((((((..(((...(((......((((((.(.(((((....))))).).)))))).....))).)))..)))))) ( -45.50) >DroSim_CAF1 92349 120 + 1 GUUUGUGCGAAGCCAUGGCCGUCCAGACGCCACUUGUCCUGAUAGUGGUGGUGAUGUACUCAAAUAUCGGUGGCACCCUCACUCCAGUUGGAGAUCCACCCAAUGUGAUCAUUGCCACCA ....(((((..(((((.((..((..((((.....))))..))..)).)))))..))))).........(((((((...((((....(((((........)))))))))....))))))). ( -41.60) >DroEre_CAF1 92998 120 + 1 GUUUAUGCGAAGCCAUGGCCGUCCGGACACCACUCGUCCUGAUAGCUGUGGUGAUGUACUCCAAUAUCGGUGGCACCCUCACACCGGUGGGUGAUCCACCCAAUGUGAUCAUUGCCACCA .....((((..((((((((..((.((((.......)))).))..))))))))..))))..........(((((((...(((((....((((((...)))))).)))))....))))))). ( -50.80) >DroYak_CAF1 91434 120 + 1 GCUUAUGCGAGGCCAUGUCCGUCCAGACACCACUUGUCCUCAUAGCUGUGGUAAUGUACUCCAAUAUCGGUGGCACCCUCACUCCAGUGGGCGAUCCACCCAAUGUGAUCAUUGCCACCA ((....))(((..((((((......)))(((((..((.......)).))))).)))..))).......(((((((..(((((....))))).((((((.....)).))))..))))))). ( -37.50) >consensus GUUUGUGCGAAGCCAUGGCCGUCCAGACGCCACUUGUCCUGAUAGUGGUGGUGAUGUACUCAAAUAUCGGUGGCACCCUCACUCCAGUGGGAGAUCCACCCAAUGUGAUCAUUGCCACCA ....(((((..(((((.((..((..((((.....))))..))..)).)))))..))))).........(((((((..(((((....))))).((((((.....)).))))..))))))). (-35.14 = -36.34 + 1.20)

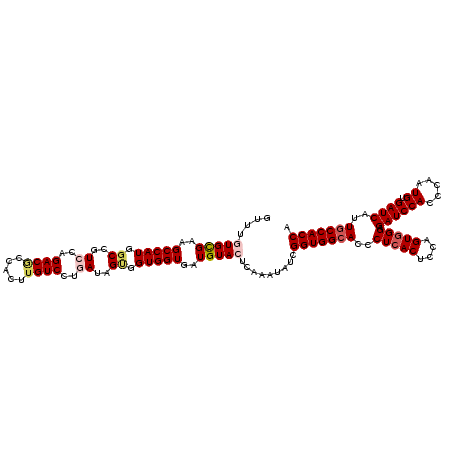
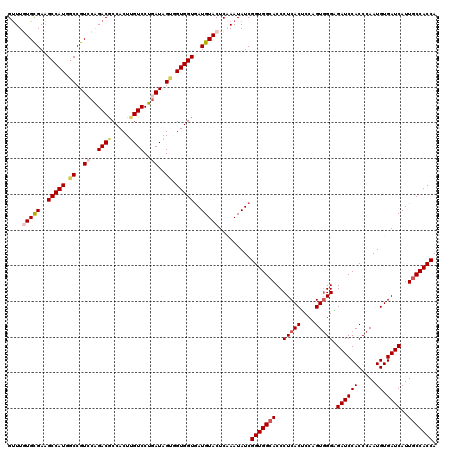
| Location | 12,970,179 – 12,970,299 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.00 |
| Mean single sequence MFE | -44.76 |
| Consensus MFE | -36.92 |
| Energy contribution | -36.52 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.63 |
| SVM RNA-class probability | 0.995883 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12970179 120 + 23771897 GAUAGUGGUGGUGAUGUACUCAAAUAUCGGUGGCACCCUCACGCCAGUGGGUGAUCCACCCAAUGUGAUCAUUGCCACCAAUUCGGAAGUGAUCUCCGCCGGAGUGGAUUUCCUCGAUUU ((...((((((..(((...(((......((((((((((.(......).)))))..))))).....))).)))..))))))..))((((((...((((...))))...))))))....... ( -47.00) >DroSec_CAF1 87523 120 + 1 GAUAGUGGUGGUGAUGUACUCAAAUAUCGGUGGAACCCUCACUCCAGUGGGAGAUCCACCCAAUGUGAUCAUUGCCACCAAUGCGGAGGUGGUCGCCGCCGGAGUGGAUUUCCUCGAUUU .....((((((..(((...(((......((((((.(.(((((....))))).).)))))).....))).)))..))))))...(((.(((((...)))))((((.....))))))).... ( -46.60) >DroSim_CAF1 92389 120 + 1 GAUAGUGGUGGUGAUGUACUCAAAUAUCGGUGGCACCCUCACUCCAGUUGGAGAUCCACCCAAUGUGAUCAUUGCCACCAAUGUGGAGGUGGUCGCCGCCGGAGUGGAUUUCCUCGAUUU ...(((((.((((...((((........)))).)))).)))))......((((((((((((...((((((((..((((....))))..))))))))....)).))))))))))....... ( -47.50) >DroEre_CAF1 93038 120 + 1 GAUAGCUGUGGUGAUGUACUCCAAUAUCGGUGGCACCCUCACACCGGUGGGUGAUCCACCCAAUGUGAUCAUUGCCACCAAUCCAGAGGUGAUCGCCAAAGGAGUGGAUUUCGUUGUAUU .(((((.(....(((.((((((......(((((((...(((((....((((((...)))))).)))))....)))))))........(((....)))...)))))).))).))))))... ( -43.30) >DroYak_CAF1 91474 120 + 1 CAUAGCUGUGGUAAUGUACUCCAAUAUCGGUGGCACCCUCACUCCAGUGGGCGAUCCACCCAAUGUGAUCAUUGCCACCAAUACGGAAGUGAUUGCCAAAGGAGUGGAUUUUCUCAAUUU ......((.((.(((.((((((......(((((((..(((((....))))).((((((.....)).))))..)))))))..(((....))).........)))))).))).)).)).... ( -39.40) >consensus GAUAGUGGUGGUGAUGUACUCAAAUAUCGGUGGCACCCUCACUCCAGUGGGAGAUCCACCCAAUGUGAUCAUUGCCACCAAUGCGGAGGUGAUCGCCGCCGGAGUGGAUUUCCUCGAUUU .....((((((((((.((((....(((.(((((((..(((((....))))).((((((.....)).))))..))))))).)))....))))))))))))))(((........)))..... (-36.92 = -36.52 + -0.40)

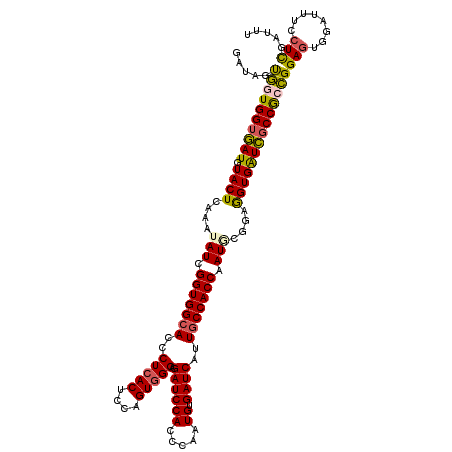
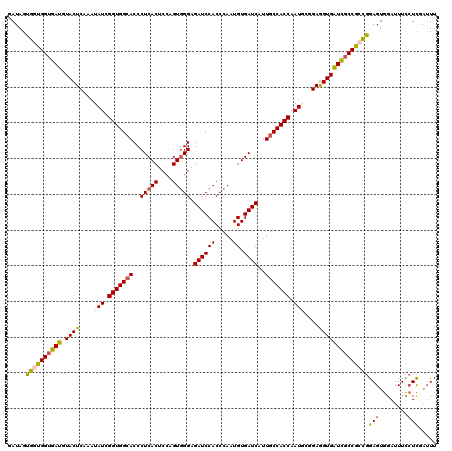
| Location | 12,970,179 – 12,970,299 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.00 |
| Mean single sequence MFE | -42.92 |
| Consensus MFE | -33.82 |
| Energy contribution | -34.22 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.79 |
| SVM decision value | 2.34 |
| SVM RNA-class probability | 0.992590 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12970179 120 - 23771897 AAAUCGAGGAAAUCCACUCCGGCGGAGAUCACUUCCGAAUUGGUGGCAAUGAUCACAUUGGGUGGAUCACCCACUGGCGUGAGGGUGCCACCGAUAUUUGAGUACAUCACCACCACUAUC .....(((........))).((.((.(((.((((....((((((((((....((((..(.(((((.....))))).).))))...))))))))))....))))..))).)).))...... ( -40.70) >DroSec_CAF1 87523 120 - 1 AAAUCGAGGAAAUCCACUCCGGCGGCGACCACCUCCGCAUUGGUGGCAAUGAUCACAUUGGGUGGAUCUCCCACUGGAGUGAGGGUUCCACCGAUAUUUGAGUACAUCACCACCACUAUC ..((((.((((..(((((((((.((.(((((((.((((....))))(((((....)))))))))).))..)).)))))))).)..))))..))))...(((.....)))........... ( -43.00) >DroSim_CAF1 92389 120 - 1 AAAUCGAGGAAAUCCACUCCGGCGGCGACCACCUCCACAUUGGUGGCAAUGAUCACAUUGGGUGGAUCUCCAACUGGAGUGAGGGUGCCACCGAUAUUUGAGUACAUCACCACCACUAUC .....(((........))).((.((.((....(((...((((((((((....((((.(..(.(((....))).)..).))))...))))))))))....)))....)).)).))...... ( -40.50) >DroEre_CAF1 93038 120 - 1 AAUACAACGAAAUCCACUCCUUUGGCGAUCACCUCUGGAUUGGUGGCAAUGAUCACAUUGGGUGGAUCACCCACCGGUGUGAGGGUGCCACCGAUAUUGGAGUACAUCACCACAGCUAUC ...................((.(((.(((...(((..(((((((((((....(((((((((.(((.....))))))))))))...)))))))))).)..)))...))).))).))..... ( -46.60) >DroYak_CAF1 91474 120 - 1 AAAUUGAGAAAAUCCACUCCUUUGGCAAUCACUUCCGUAUUGGUGGCAAUGAUCACAUUGGGUGGAUCGCCCACUGGAGUGAGGGUGCCACCGAUAUUGGAGUACAUUACCACAGCUAUG .....(((........)))((.(((.(((.(((((.((((((((((((....((((.((.(((((.....))))).))))))...)))))))))))).)))))..))).))).))..... ( -43.80) >consensus AAAUCGAGGAAAUCCACUCCGGCGGCGAUCACCUCCGCAUUGGUGGCAAUGAUCACAUUGGGUGGAUCACCCACUGGAGUGAGGGUGCCACCGAUAUUUGAGUACAUCACCACCACUAUC ....................((.((.(((...(((...((((((((((....((((.((.(((((.....))))).))))))...))))))))))....)))...))).)).))...... (-33.82 = -34.22 + 0.40)


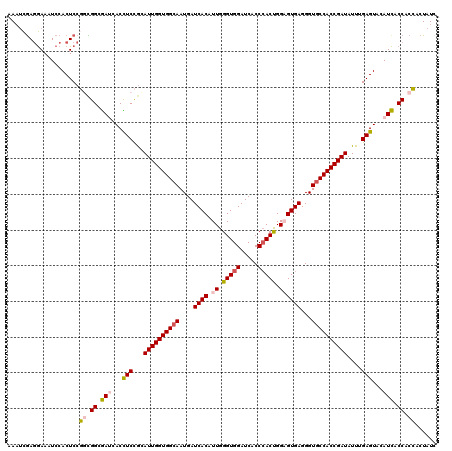
| Location | 12,970,219 – 12,970,339 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.83 |
| Mean single sequence MFE | -40.18 |
| Consensus MFE | -32.56 |
| Energy contribution | -32.52 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.81 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.522122 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12970219 120 + 23771897 ACGCCAGUGGGUGAUCCACCCAAUGUGAUCAUUGCCACCAAUUCGGAAGUGAUCUCCGCCGGAGUGGAUUUCCUCGAUUUCACCAUUCACAUGUUGCCUGGCGUUUUAUUGGCGGCUGUA .((((((((((((...))))).....(((((((.((........)).)))))))..((((((.(((((..........)))))(((....)))....))))))....)))))))...... ( -38.20) >DroSec_CAF1 87563 120 + 1 ACUCCAGUGGGAGAUCCACCCAAUGUGAUCAUUGCCACCAAUGCGGAGGUGGUCGCCGCCGGAGUGGAUUUCCUCGAUUUCACCAUUCACAUGUUGCCUGGCGUUAUAAUAGCGGCUGUA ....(((((((((((((((((...((((((((..((........))..))))))))....)).)))))))))))........(((..((.....))..)))(((((...))))))))).. ( -39.60) >DroSim_CAF1 92429 120 + 1 ACUCCAGUUGGAGAUCCACCCAAUGUGAUCAUUGCCACCAAUGUGGAGGUGGUCGCCGCCGGAGUGGAUUUCCUCGAUUUCACCAUUCACAUGUUGCCUGGCGUUGUAAUGGCGGCUGUA ....(((((((((((((((((...((((((((..((((....))))..))))))))....)).)))))))))).........(((((.(((((((....)))).)))))))).))))).. ( -47.00) >DroEre_CAF1 93078 120 + 1 ACACCGGUGGGUGAUCCACCCAAUGUGAUCAUUGCCACCAAUCCAGAGGUGAUCGCCAAAGGAGUGGAUUUCGUUGUAUUCACCCUGCACAUGCUGCCAGGUGUUUUAAUGGCGGCUGUA .(((((((((((((((((.....)).))))))..)))))..(....)))))...((...(((.((((((........)))))))))))(((.(((((((..........)))))))))). ( -41.20) >DroYak_CAF1 91514 120 + 1 ACUCCAGUGGGCGAUCCACCCAAUGUGAUCAUUGCCACCAAUACGGAAGUGAUUGCCAAAGGAGUGGAUUUUCUCAAUUUCACAAUUCACAUGCUGCCUGGAGUUUUAAUGGCUGGUGUG (((((((..((((((((((((...(..((((((.((........)).))))))..)....)).))))))).....((((....)))).....)))..)))))))................ ( -34.90) >consensus ACUCCAGUGGGAGAUCCACCCAAUGUGAUCAUUGCCACCAAUGCGGAGGUGAUCGCCGCCGGAGUGGAUUUCCUCGAUUUCACCAUUCACAUGUUGCCUGGCGUUUUAAUGGCGGCUGUA .((((....))))((((((((...(((((((((.((........)).)))))))))....)).))))))...................(((.((((((............))))))))). (-32.56 = -32.52 + -0.04)

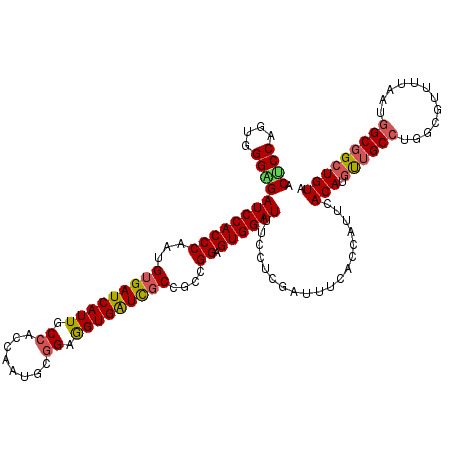
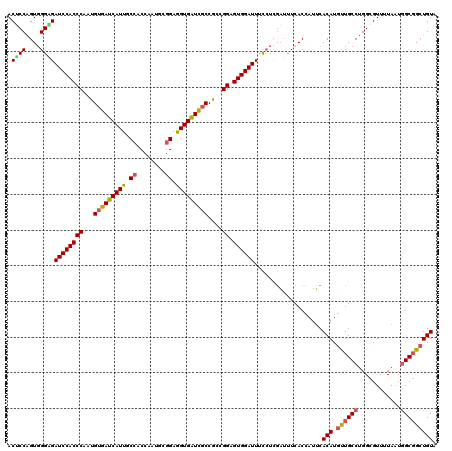
| Location | 12,970,219 – 12,970,339 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.83 |
| Mean single sequence MFE | -40.32 |
| Consensus MFE | -27.06 |
| Energy contribution | -28.06 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.616105 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12970219 120 - 23771897 UACAGCCGCCAAUAAAACGCCAGGCAACAUGUGAAUGGUGAAAUCGAGGAAAUCCACUCCGGCGGAGAUCACUUCCGAAUUGGUGGCAAUGAUCACAUUGGGUGGAUCACCCACUGGCGU ....(((((((((....(((((..((.....))..)))))...(((.((....)).((((...))))........))))))))))))........((.((((((...)))))).)).... ( -41.10) >DroSec_CAF1 87563 120 - 1 UACAGCCGCUAUUAUAACGCCAGGCAACAUGUGAAUGGUGAAAUCGAGGAAAUCCACUCCGGCGGCGACCACCUCCGCAUUGGUGGCAAUGAUCACAUUGGGUGGAUCUCCCACUGGAGU ....((((((.......(((((..((.....))..))))).....(((........))).))))))..(((((.((((....))))(((((....))))))))))..((((....)))). ( -42.50) >DroSim_CAF1 92429 120 - 1 UACAGCCGCCAUUACAACGCCAGGCAACAUGUGAAUGGUGAAAUCGAGGAAAUCCACUCCGGCGGCGACCACCUCCACAUUGGUGGCAAUGAUCACAUUGGGUGGAUCUCCAACUGGAGU ....((((((.......(((((..((.....))..))))).....(((........))).))))))..(((((.((((....))))(((((....))))))))))..((((....)))). ( -46.60) >DroEre_CAF1 93078 120 - 1 UACAGCCGCCAUUAAAACACCUGGCAGCAUGUGCAGGGUGAAUACAACGAAAUCCACUCCUUUGGCGAUCACCUCUGGAUUGGUGGCAAUGAUCACAUUGGGUGGAUCACCCACCGGUGU ....(((((((......(((((.(((.....))).))))).................(((...((......))...))).)))))))......(((..((((((...))))))...))). ( -38.60) >DroYak_CAF1 91514 120 - 1 CACACCAGCCAUUAAAACUCCAGGCAGCAUGUGAAUUGUGAAAUUGAGAAAAUCCACUCCUUUGGCAAUCACUUCCGUAUUGGUGGCAAUGAUCACAUUGGGUGGAUCGCCCACUGGAGU ................(((((((.....(((((((((((.(((..(((........))).))).)))))((((........)))).......))))))((((((...))))))))))))) ( -32.80) >consensus UACAGCCGCCAUUAAAACGCCAGGCAACAUGUGAAUGGUGAAAUCGAGGAAAUCCACUCCGGCGGCGAUCACCUCCGCAUUGGUGGCAAUGAUCACAUUGGGUGGAUCACCCACUGGAGU ....((((((.......(((((..((.....))..))))).....(((........))).))))))..(((((........)))))(((((....)))))(((((.....)))))..... (-27.06 = -28.06 + 1.00)

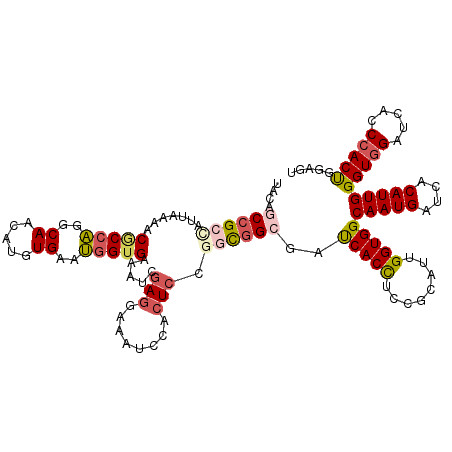
| Location | 12,970,259 – 12,970,379 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.67 |
| Mean single sequence MFE | -31.47 |
| Consensus MFE | -21.64 |
| Energy contribution | -21.48 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.592266 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12970259 120 - 23771897 AAGAUUUCCUCAUGGUCACAUACAUGACAGCAUAGCCCGCUACAGCCGCCAAUAAAACGCCAGGCAACAUGUGAAUGGUGAAAUCGAGGAAAUCCACUCCGGCGGAGAUCACUUCCGAAU ..(((((((((....((((.((((((..(((.......)))...(((...............)))..))))))....))))....)))))))))........(((((.....)))))... ( -34.26) >DroSec_CAF1 87603 120 - 1 AAGAUUUCCUCAUGGUCACAUACAUGACAGCAUAGCCCGCUACAGCCGCUAUUAUAACGCCAGGCAACAUGUGAAUGGUGAAAUCGAGGAAAUCCACUCCGGCGGCGACCACCUCCGCAU ..(((((((((....((((.((((((..(((.......)))...(((((.........))..)))..))))))....))))....))))))))).......((((.(.....).)))).. ( -36.40) >DroSim_CAF1 92469 120 - 1 AAGAUUUCCUCAUGGUCACAUACAUGACAGCAUAGCCCGCUACAGCCGCCAUUACAACGCCAGGCAACAUGUGAAUGGUGAAAUCGAGGAAAUCCACUCCGGCGGCGACCACCUCCACAU ............(((((...........(((.......)))...((((((.......(((((..((.....))..))))).....(((........))).)))))))))))......... ( -35.00) >DroEre_CAF1 93118 120 - 1 AAGAUUUUCUCAUAGUCAGAUAAAUGACACAAUAACCCGAUACAGCCGCCAUUAAAACACCUGGCAGCAUGUGCAGGGUGAAUACAACGAAAUCCACUCCUUUGGCGAUCACCUCUGGAU ..((((((......((((......))))......((((..((((((.((((..........)))).)).))))..)))).........))))))...(((...((......))...))). ( -29.20) >DroYak_CAF1 91554 120 - 1 GGGAUUUCCUCAUGGUCAGAUAUAUGACACCAUAACCCGCCACACCAGCCAUUAAAACUCCAGGCAGCAUGUGAAUUGUGAAAUUGAGAAAAUCCACUCCUUUGGCAAUCACUUCCGUAU .((((((.((((..((((......))))..(((((..(((.......(((............))).....)))..)))))....)))).))))))......................... ( -22.50) >consensus AAGAUUUCCUCAUGGUCACAUACAUGACAGCAUAGCCCGCUACAGCCGCCAUUAAAACGCCAGGCAACAUGUGAAUGGUGAAAUCGAGGAAAUCCACUCCGGCGGCGAUCACCUCCGCAU ..(((((((((...((((......))))......(((...((((...(((............)))....))))...)))......))))))))).......((((.(.....).)))).. (-21.64 = -21.48 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:24:23 2006