
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,966,803 – 12,966,963 |
| Length | 160 |
| Max. P | 0.989788 |

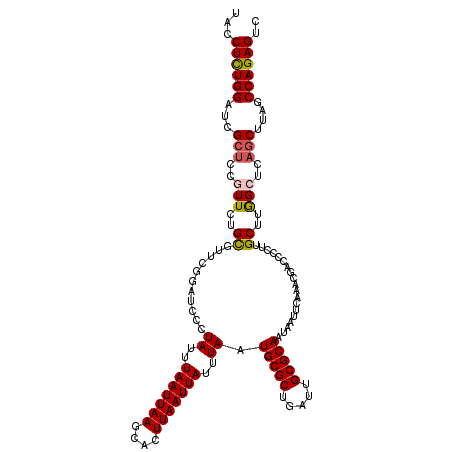
| Location | 12,966,803 – 12,966,923 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.33 |
| Mean single sequence MFE | -30.30 |
| Consensus MFE | -23.88 |
| Energy contribution | -24.04 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.79 |
| SVM decision value | 2.18 |
| SVM RNA-class probability | 0.989788 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12966803 120 + 23771897 UACCUCUGGAUCGCUUCGUUCUGUGUUCGGAUCCCUAUUUAAUUAAGCACUUAAUUAUUUAAUGCGCUGAUUGCGCAAUAAUUCAAACGACCCCUUGCUUGGCUCAGCUUAGCCAGAGUC ...((((((...(((..(((..(((...((.((..((..(((((((....)))))))..)).(((((.....)))))...........)).))..)))..)))..)))....)))))).. ( -29.20) >DroSec_CAF1 84042 120 + 1 UACCUCUGGAUCGCUCCAUUCUGCGUUCGUAUCCCUAUUUAAUUAAGCACUUAAUUAUUUAAUGCGCUGAUUGCGCAAUAAUUCAAACGACCCUUUGCUUGGCUCAGCUUAGCCAGAGUC ...((((((...((((((....(((.((((.....((..(((((((....)))))))..)).(((((.....))))).........)))).....))).)))...)))....)))))).. ( -30.30) >DroSim_CAF1 88944 120 + 1 UACCUCUGGAUCGCUCCAUUCUGCGUUCGUAUCCCUAUUUAAUUAAGCACUUAAUUAUUUAAUGCGCUGAUUGCGCAAUAAUUCAAACGACCCCUUGCUUGGCUCAGCUUAGCCAGAGUC ...((((((...((((((....(((.((((.....((..(((((((....)))))))..)).(((((.....))))).........)))).....))).)))...)))....)))))).. ( -30.30) >DroEre_CAF1 89863 120 + 1 UACCUUUGGAUCGCAGCGUUCUGUUUCCUGCUUCCUAUUUAAUUAAGCACUUAAUUAUUUAAUGCGCUGAUUGCGCAAUAUUUCAAACGACUCCAUGCUUAGCUCAACUUAGCCAGAGUC .......(((..((((.(........))))).)))((..(((((((....)))))))..)).(((((.....)))))...........(((((...(((.((.....)).)))..))))) ( -27.20) >DroYak_CAF1 87984 120 + 1 UACCUCUGGUUCGCUGCGUUCUGCUUCCUGCUCCCUAUUUAAUUAAGCACUUAAUUAUUUAAUGCGCUGAUUGCGCAAUAUUUCAACCGACUCCUUGCUUAGCUCAGCUCAGCCAGAGUC ...((((((((.((((.(((..((...........((..(((((((....)))))))..)).(((((.....)))))...................))..))).))))..)))))))).. ( -34.50) >consensus UACCUCUGGAUCGCUCCGUUCUGCGUUCGGAUCCCUAUUUAAUUAAGCACUUAAUUAUUUAAUGCGCUGAUUGCGCAAUAAUUCAAACGACCCCUUGCUUGGCUCAGCUUAGCCAGAGUC ...((((((...(((..(((..((...........((..(((((((....)))))))..)).(((((.....)))))...................))..)))..)))....)))))).. (-23.88 = -24.04 + 0.16)


| Location | 12,966,803 – 12,966,923 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.33 |
| Mean single sequence MFE | -33.76 |
| Consensus MFE | -26.44 |
| Energy contribution | -27.72 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.960170 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12966803 120 - 23771897 GACUCUGGCUAAGCUGAGCCAAGCAAGGGGUCGUUUGAAUUAUUGCGCAAUCAGCGCAUUAAAUAAUUAAGUGCUUAAUUAAAUAGGGAUCCGAACACAGAACGAAGCGAUCCAGAGGUA ..((((((((......)))).......(((((((((...(((.(((((.....))))).))).(((((((....))))))).......................)))))))))))))... ( -34.10) >DroSec_CAF1 84042 120 - 1 GACUCUGGCUAAGCUGAGCCAAGCAAAGGGUCGUUUGAAUUAUUGCGCAAUCAGCGCAUUAAAUAAUUAAGUGCUUAAUUAAAUAGGGAUACGAACGCAGAAUGGAGCGAUCCAGAGGUA ..((((((((......)))).......((((((((((..(((.(((((.....))))).))).(((((((....)))))))..........)))))((........)))))))))))... ( -35.40) >DroSim_CAF1 88944 120 - 1 GACUCUGGCUAAGCUGAGCCAAGCAAGGGGUCGUUUGAAUUAUUGCGCAAUCAGCGCAUUAAAUAAUUAAGUGCUUAAUUAAAUAGGGAUACGAACGCAGAAUGGAGCGAUCCAGAGGUA ..((((((((......)))).......((((((((((..(((.(((((.....))))).))).(((((((....)))))))..........)))))((........)))))))))))... ( -35.20) >DroEre_CAF1 89863 120 - 1 GACUCUGGCUAAGUUGAGCUAAGCAUGGAGUCGUUUGAAAUAUUGCGCAAUCAGCGCAUUAAAUAAUUAAGUGCUUAAUUAAAUAGGAAGCAGGAAACAGAACGCUGCGAUCCAAAGGUA ((((((((((.((.....)).))).)))))))........((.(((((.....))))).))..(((((((....)))))))....(((.(((((........).))))..)))....... ( -32.30) >DroYak_CAF1 87984 120 - 1 GACUCUGGCUGAGCUGAGCUAAGCAAGGAGUCGGUUGAAAUAUUGCGCAAUCAGCGCAUUAAAUAAUUAAGUGCUUAAUUAAAUAGGGAGCAGGAAGCAGAACGCAGCGAACCAGAGGUA ..((((((.(..((((...((((((...(((...((((.....(((((.....)))))))))...)))...))))))............((.....))......))))..)))))))... ( -31.80) >consensus GACUCUGGCUAAGCUGAGCCAAGCAAGGGGUCGUUUGAAUUAUUGCGCAAUCAGCGCAUUAAAUAAUUAAGUGCUUAAUUAAAUAGGGAUCCGAACGCAGAACGCAGCGAUCCAGAGGUA ..((((((((......)))).......(((((((((....((.(((((.....))))).))..(((((((....))))))).......................)))))))))))))... (-26.44 = -27.72 + 1.28)


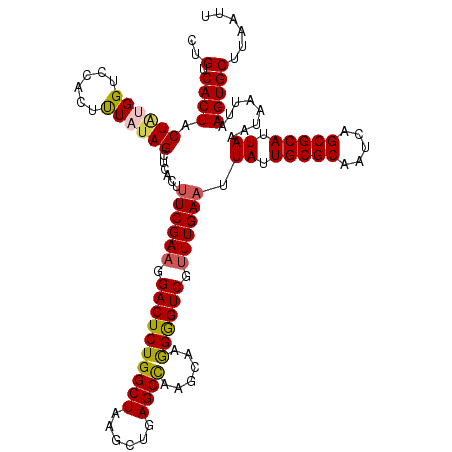
| Location | 12,966,843 – 12,966,963 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.17 |
| Mean single sequence MFE | -32.24 |
| Consensus MFE | -28.72 |
| Energy contribution | -28.80 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.91 |
| SVM RNA-class probability | 0.982322 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12966843 120 - 23771897 CUGUCACUACUAUGGUCCACUUUAUAGCUAACUUUCGAAGGACUCUGGCUAAGCUGAGCCAAGCAAGGGGUCGUUUGAAUUAUUGCGCAAUCAGCGCAUUAAAUAAUUAAGUGCUUAAUU ..(.((((.((((((......))))))......((((((.((((((((((......))))......)))))).))))))(((.(((((.....))))).))).......)))))...... ( -31.70) >DroSec_CAF1 84082 120 - 1 CUGUCACUACUAUGGUCCACUCUAUAGCUCACUUUCGAAGGACUCUGGCUAAGCUGAGCCAAGCAAAGGGUCGUUUGAAUUAUUGCGCAAUCAGCGCAUUAAAUAAUUAAGUGCUUAAUU ..(.((((.((((((......))))))......((((((.((((((((((......))))......)))))).))))))(((.(((((.....))))).))).......)))))...... ( -33.70) >DroSim_CAF1 88984 120 - 1 CUGUCACUACUAUGGUCCACUUUAUAGCUCACUUUCGAAGGACUCUGGCUAAGCUGAGCCAAGCAAGGGGUCGUUUGAAUUAUUGCGCAAUCAGCGCAUUAAAUAAUUAAGUGCUUAAUU ..(.((((.((((((......))))))......((((((.((((((((((......))))......)))))).))))))(((.(((((.....))))).))).......)))))...... ( -31.70) >DroEre_CAF1 89903 120 - 1 GUGCCACUACUGAGGUCCACUUUAUAGCUCAAAUUCGAAGGACUCUGGCUAAGUUGAGCUAAGCAUGGAGUCGUUUGAAAUAUUGCGCAAUCAGCGCAUUAAAUAAUUAAGUGCUUAAUU (..((.(....).))..).......(((.(...((((((.((((((((((.((.....)).))).))))))).)))))).((.(((((.....))))).)).........).)))..... ( -33.90) >DroYak_CAF1 88024 120 - 1 CUGUCACUACUUGGGUCCACUUUAUAGCUCACAAUCGAAGGACUCUGGCUGAGCUGAGCUAAGCAAGGAGUCGGUUGAAAUAUUGCGCAAUCAGCGCAUUAAAUAAUUAAGUGCUUAAUU ..(.((((...(((((..........)))))...((((..((((((.(((.(((...))).)))..))))))..))))..((.(((((.....))))).))........)))))...... ( -30.20) >consensus CUGUCACUACUAUGGUCCACUUUAUAGCUCACUUUCGAAGGACUCUGGCUAAGCUGAGCCAAGCAAGGGGUCGUUUGAAUUAUUGCGCAAUCAGCGCAUUAAAUAAUUAAGUGCUUAAUU ..(.((((.((((((......))))))......((((((.((((((((((......))))......)))))).)))))).((.(((((.....))))).))........)))))...... (-28.72 = -28.80 + 0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:24:13 2006