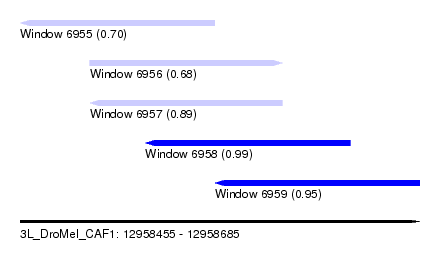
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,958,455 – 12,958,685 |
| Length | 230 |
| Max. P | 0.991962 |

| Location | 12,958,455 – 12,958,567 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.71 |
| Mean single sequence MFE | -32.10 |
| Consensus MFE | -23.66 |
| Energy contribution | -23.50 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.703326 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12958455 112 - 23771897 CGCUGAGGGUUUGGAAGAAGUCAAGCCCUCCUUAUUGCACCAGCAAUG-------UGUGUGUGCGGCAAAACUU-UAGUUGAGUUUCGGCUAAGUUAUUCGAAAUUUUAGUAAUUGAUAU (((.((((((((((......))))))))))......((((((....))-------.))))..)))....(((((-.(((((.....))))))))))........................ ( -31.00) >DroSec_CAF1 75678 113 - 1 CGCUGAGGGUUUGGAAAAAGUCAAGCCCUCCUUAUUGCACCAGCAAUU-------UGUGGGUGCGGGAAAACUUUUGGUUGAGUUUCGGCUAAGUUAUACGAAAUUUUAGUAAUUUAUAU .(((((((((((((......))))))))((((....(((((.((....-------.)).))))))))).(((((......))))))))))((((((((...........))))))))... ( -33.20) >DroSim_CAF1 80424 113 - 1 CGCUGAGGGUUUGGAAAAAGUCAAACCCUCCCUAUUGCACCAGCAAUG-------UGUGGGUGCGGGAAAACUUUUGGUUGAGUUUCGGCUAAGUUAUACGAAAUUUUAGUAAUUGAUAU ....((((((((((......))))))))))(((...(((((.((....-------.)).))))))))....(..(((.(.((((((((...........)))))))).).)))..).... ( -33.90) >DroEre_CAF1 81728 112 - 1 CUUCGAGGGCUUGGGAAAAGUCAGGCCCUCCUUAUUACAGCAGUAAUG-------UAUGUGGGCGGGAAAACUU-UGGUUGAGUUUCGUCUAAGUUAUCCAACAUUUUAGUAAUUGAUAU ....((((((((((......))))))))))...(((((.(((....))-------)...((((((..(.(((..-..)))...)..)))))).................)))))...... ( -32.40) >DroYak_CAF1 79555 119 - 1 CUCCGAGGGUUUGGAAAAAGUCAAGCCCUCCGUAUUACAUCAGCAAUGGUACGAGUAUAUGUGCGUGAAAACUU-UGGUUGCGUUUCGACUAAGUUAUCCAACAUUUUAGUAAUUGAUAU ....((((((((((......))))))))))((.(((((....(((...(((....)))...)))(.((.(((((-.(((((.....)))))))))).))).........))))))).... ( -30.00) >consensus CGCUGAGGGUUUGGAAAAAGUCAAGCCCUCCUUAUUGCACCAGCAAUG_______UGUGUGUGCGGGAAAACUU_UGGUUGAGUUUCGGCUAAGUUAUACGAAAUUUUAGUAAUUGAUAU (((.((((((((((......))))))))))..((((((....))))))..............)))....((((..((((((.....))))))))))........................ (-23.66 = -23.50 + -0.16)



| Location | 12,958,495 – 12,958,606 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.64 |
| Mean single sequence MFE | -32.92 |
| Consensus MFE | -24.44 |
| Energy contribution | -25.08 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.684647 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12958495 111 + 23771897 AACUA-AAGUUUUGCCGCACACACA-------CAUUGCUGGUGCAAUAAGGAGGGCUUGACUUCUUCCAAACCCUCAGCGAGGGCGACAAUGACUUUGUGUAAAUAUA-GGGGGAGAAUU .((((-((((((((((((((.((..-------......))))))......(((((.(((........))).)))))......)))))....))))))).)).......-........... ( -27.80) >DroSec_CAF1 75718 112 + 1 AACCAAAAGUUUUCCCGCACCCACA-------AAUUGCUGGUGCAAUAAGGAGGGCUUGACUUUUUCCAAACCCUCAGCGAGGGCGACAAUGACUUUGUGUAAAUAUA-GGGGGAGAAUU .......(((((((((.(.(((...-------.(((((....)))))...(((((.(((........))).))))).....)))..((((.....)))).........-).))))))))) ( -33.70) >DroSim_CAF1 80464 112 + 1 AACCAAAAGUUUUCCCGCACCCACA-------CAUUGCUGGUGCAAUAGGGAGGGUUUGACUUUUUCCAAACCCUCAGCGAGGGCGACAAUGACUUUGUGUAAAUAUA-GGGGGAGAAUU .......(((((((((.(.(((.(.-------.(((((....)))))..)(((((((((........))))))))).....)))..((((.....)))).........-).))))))))) ( -38.20) >DroEre_CAF1 81768 112 + 1 AACCA-AAGUUUUCCCGCCCACAUA-------CAUUACUGCUGUAAUAAGGAGGGCCUGACUUUUCCCAAGCCCUCGAAGAGGGCGACAAUGACUUUGUGUAAAUAUAUAGGGGAGAAUU .....-.(((((((((((((.....-------.(((((....)))))...((((((.((........)).)))))).....)))).((((.....))))............))))))))) ( -36.80) >DroYak_CAF1 79595 118 + 1 AACCA-AAGUUUUCACGCACAUAUACUCGUACCAUUGCUGAUGUAAUACGGAGGGCUUGACUUUUUCCAAACCCUCGGAGAGGACGACAAUGACUUUGUGUAAAUAUA-GGGGGAGAAUU .....-.(((((((.....(((....((((.(((((((....))))).(.(((((.(((........))).))))).)...))))))..))).(((((((....))))-))).))))))) ( -28.10) >consensus AACCA_AAGUUUUCCCGCACACACA_______CAUUGCUGGUGCAAUAAGGAGGGCUUGACUUUUUCCAAACCCUCAGCGAGGGCGACAAUGACUUUGUGUAAAUAUA_GGGGGAGAAUU .......(((((((((.(...............(((((....)))))...(((((.(((........))).))))).((((((..........))))))..........).))))))))) (-24.44 = -25.08 + 0.64)



| Location | 12,958,495 – 12,958,606 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.64 |
| Mean single sequence MFE | -34.11 |
| Consensus MFE | -24.10 |
| Energy contribution | -24.38 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.885632 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12958495 111 - 23771897 AAUUCUCCCCC-UAUAUUUACACAAAGUCAUUGUCGCCCUCGCUGAGGGUUUGGAAGAAGUCAAGCCCUCCUUAUUGCACCAGCAAUG-------UGUGUGUGCGGCAAAACUU-UAGUU ...........-...........(((((..(((((((.(.(((.((((((((((......))))))))))..((((((....))))))-------.))).).))))))).))))-).... ( -35.20) >DroSec_CAF1 75718 112 - 1 AAUUCUCCCCC-UAUAUUUACACAAAGUCAUUGUCGCCCUCGCUGAGGGUUUGGAAAAAGUCAAGCCCUCCUUAUUGCACCAGCAAUU-------UGUGGGUGCGGGAAAACUUUUGGUU ...........-........((.(((((..((.((((.(((((.((((((((((......))))))))))...(((((....))))).-------.))))).)))).)).)))))))... ( -33.30) >DroSim_CAF1 80464 112 - 1 AAUUCUCCCCC-UAUAUUUACACAAAGUCAUUGUCGCCCUCGCUGAGGGUUUGGAAAAAGUCAAACCCUCCCUAUUGCACCAGCAAUG-------UGUGGGUGCGGGAAAACUUUUGGUU .....((((..-.........((((.....))))(((((.(((.((((((((((......))))))))))...(((((....))))))-------)).))))).))))............ ( -35.70) >DroEre_CAF1 81768 112 - 1 AAUUCUCCCCUAUAUAUUUACACAAAGUCAUUGUCGCCCUCUUCGAGGGCUUGGGAAAAGUCAGGCCCUCCUUAUUACAGCAGUAAUG-------UAUGUGGGCGGGAAAACUU-UGGUU ......................((((((..((.((((((.....((((((((((......))))))))))......((((((....))-------).))))))))).)).))))-))... ( -38.10) >DroYak_CAF1 79595 118 - 1 AAUUCUCCCCC-UAUAUUUACACAAAGUCAUUGUCGUCCUCUCCGAGGGUUUGGAAAAAGUCAAGCCCUCCGUAUUACAUCAGCAAUGGUACGAGUAUAUGUGCGUGAAAACUU-UGGUU ...........-..........((((((((((((.(......(.((((((((((......)))))))))).)........).))))))(((((......)))))......))))-))... ( -28.24) >consensus AAUUCUCCCCC_UAUAUUUACACAAAGUCAUUGUCGCCCUCGCUGAGGGUUUGGAAAAAGUCAAGCCCUCCUUAUUGCACCAGCAAUG_______UGUGUGUGCGGGAAAACUU_UGGUU ...................((...((((..((.((((.(.(((.((((((((((......))))))))))..((((((....))))))........))).).)))).)).))))...)). (-24.10 = -24.38 + 0.28)



| Location | 12,958,527 – 12,958,645 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.45 |
| Mean single sequence MFE | -25.56 |
| Consensus MFE | -24.47 |
| Energy contribution | -24.23 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.30 |
| SVM RNA-class probability | 0.991962 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12958527 118 - 23771897 CCCAUUGCAAUAACAAAUAUUGGU-UGCUGGCACAAUUUCAAUUCUCCCCC-UAUAUUUACACAAAGUCAUUGUCGCCCUCGCUGAGGGUUUGGAAGAAGUCAAGCCCUCCUUAUUGCAC .....((((((((...........-.((.((((((((..(...........-..............)..))))).)))...)).((((((((((......)))))))))).)))))))). ( -28.01) >DroSec_CAF1 75751 119 - 1 CCCAUUGCAAUAACAAAUAUUGGUUUGCUGGCACAAUUUCAAUUCUCCCCC-UAUAUUUACACAAAGUCAUUGUCGCCCUCGCUGAGGGUUUGGAAAAAGUCAAGCCCUCCUUAUUGCAC .....((((((((.............((.((((((((..(...........-..............)..))))).)))...)).((((((((((......)))))))))).)))))))). ( -28.01) >DroSim_CAF1 80497 119 - 1 CCCAUUGCAAUAACAAAUAUUGGUUUGCUGGCACAAUUUCAAUUCUCCCCC-UAUAUUUACACAAAGUCAUUGUCGCCCUCGCUGAGGGUUUGGAAAAAGUCAAACCCUCCCUAUUGCAC .....(((((((..............((.((((((((..(...........-..............)..))))).)))...)).((((((((((......))))))))))..))))))). ( -27.41) >DroEre_CAF1 81800 119 - 1 CCCAUUGCAAUAACAAAUAUUGGC-UGCUGGCACAAUUUCAAUUCUCCCCUAUAUAUUUACACAAAGUCAUUGUCGCCCUCUUCGAGGGCUUGGGAAAAGUCAGGCCCUCCUUAUUACAG .......(((((.....))))).(-((..((((((((..(..........................)..))))).)))......((((((((((......)))))))))).......))) ( -24.07) >DroYak_CAF1 79634 118 - 1 CCCAUUGCAAUAACAAAUAUUGGU-UGCUGGCACAAUUUCAAUUCUCCCCC-UAUAUUUACACAAAGUCAUUGUCGUCCUCUCCGAGGGUUUGGAAAAAGUCAAGCCCUCCGUAUUACAU .(((..(((((...........))-))))))....................-......(((((((.....))))..........((((((((((......)))))))))).)))...... ( -20.30) >consensus CCCAUUGCAAUAACAAAUAUUGGU_UGCUGGCACAAUUUCAAUUCUCCCCC_UAUAUUUACACAAAGUCAUUGUCGCCCUCGCUGAGGGUUUGGAAAAAGUCAAGCCCUCCUUAUUGCAC .....(((((((.................((((((((..(..........................)..))))).)))......((((((((((......))))))))))..))))))). (-24.47 = -24.23 + -0.24)

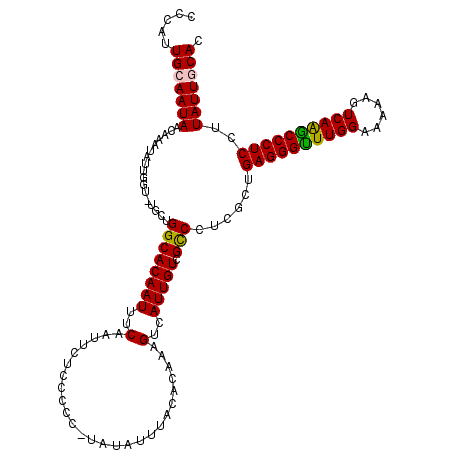

| Location | 12,958,567 – 12,958,685 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.81 |
| Mean single sequence MFE | -23.57 |
| Consensus MFE | -22.74 |
| Energy contribution | -22.18 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.952666 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12958567 118 - 23771897 AACAAAUCAGCGACAAAUGCAUGGGCGGUGCACAAACUCACCCAUUGCAAUAACAAAUAUUGGU-UGCUGGCACAAUUUCAAUUCUCCCCC-UAUAUUUACACAAAGUCAUUGUCGCCCU .........(((((((.((((((((.(((......)).).)))).)))).....((((((.((.-........................))-.))))))...........)))))))... ( -24.21) >DroSec_CAF1 75791 119 - 1 AACAAAUCAGCGACAAAUGCAUGGGCGGUGCACAAACUCACCCAUUGCAAUAACAAAUAUUGGUUUGCUGGCACAAUUUCAAUUCUCCCCC-UAUAUUUACACAAAGUCAUUGUCGCCCU .........(((((((.((((((((.(((......)).).)))).)))).........(((((((....))).))))..............-..................)))))))... ( -24.30) >DroSim_CAF1 80537 119 - 1 AACAAAUCAGCGACAAAUGCAUGGGCGGUGCACAAACUCACCCAUUGCAAUAACAAAUAUUGGUUUGCUGGCACAAUUUCAAUUCUCCCCC-UAUAUUUACACAAAGUCAUUGUCGCCCU .........(((((((.((((((((.(((......)).).)))).)))).........(((((((....))).))))..............-..................)))))))... ( -24.30) >DroEre_CAF1 81840 119 - 1 AACAAAUCAGCGACAAAUGCAUGGGCGGUGCACGAAUUCACCCAUUGCAAUAACAAAUAUUGGC-UGCUGGCACAAUUUCAAUUCUCCCCUAUAUAUUUACACAAAGUCAUUGUCGCCCU .........(((((((.((((((((.((.........)).)))).))))...........((((-(..((................................)).))))))))))))... ( -24.35) >DroYak_CAF1 79674 118 - 1 AACAAAUCAGCGACAAAUGCAUGGGCGGUGCACGAAUACACCCAUUGCAAUAACAAAUAUUGGU-UGCUGGCACAAUUUCAAUUCUCCCCC-UAUAUUUACACAAAGUCAUUGUCGUCCU ......((((((((...((((((((..(((......))).)))).))))....((.....))))-))))))....................-.......((((((.....)))).))... ( -20.70) >consensus AACAAAUCAGCGACAAAUGCAUGGGCGGUGCACAAACUCACCCAUUGCAAUAACAAAUAUUGGU_UGCUGGCACAAUUUCAAUUCUCCCCC_UAUAUUUACACAAAGUCAUUGUCGCCCU .........(((((((.((((((((.(((......)).).)))).)))).........((((((......)).)))).................................)))))))... (-22.74 = -22.18 + -0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:23:54 2006