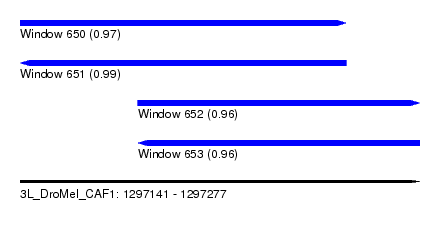
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,297,141 – 1,297,277 |
| Length | 136 |
| Max. P | 0.989997 |

| Location | 1,297,141 – 1,297,252 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 87.65 |
| Mean single sequence MFE | -36.81 |
| Consensus MFE | -28.67 |
| Energy contribution | -29.27 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.972447 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1297141 111 + 23771897 UCAAAGCGGAUCUUAAAAGUCCGCAAGCGAUGCAGCCCUAGCUUAAGCCCAUUGUCAGGGCUGCCAACGUGGGCAAUGUGGCCCUAUCCCAACCAGUUGGCAUCGCCACCA .....((((((.......))))))..(((((..((((((..(...........)..))))))((((((..((((......))))...........)))))))))))..... ( -42.02) >DroSec_CAF1 926 111 + 1 UAGGAGCGGAUCUUAAAAAUCCGAAAGCGAUGCAGCCCUAGCUUAAGUUAAUUUCCAGAGCUGCCAACGUGGGCAAAGUGGCCCUAUGCCAACCAGUUGGCAUCGCCACCA ..((..(((((.......)))))...(((((((.....((((((.............)))))).((((.(((......((((.....)))).))))))))))))))..)). ( -34.92) >DroSim_CAF1 4227 111 + 1 UAGAAGCGGAUCUUAAAAAUCCGCAAGCGAUGCAGCCCUAGCUUAAGUUCAUUUCCAGAGCUGCCAACGUGGGCAAAGUGGCCCUAUGCCAACCAGUUGGCAUCGCCACCA .....((((((.......))))))..(((((((.((((.......(((((.......)))))........))))....((((.....))))........)))))))..... ( -38.96) >DroEre_CAF1 902 110 + 1 UAGAAGUGGAGCUUUAAAAUUCGCAAGCGAUGCAGCCCUAGCUUGAAUCCAUUUACAGCGCUGCCAACGUGGGCAAAGUGGCUCU-UGCCAAUCAGUUGGCAUCGCCACCA ...(((((((.............(((((............)))))..)))))))...(((.(((((((...(((((........)-)))).....))))))).)))..... ( -34.06) >DroYak_CAF1 4140 110 + 1 UAAGAGCAAAUCUUAAAAAUUCGCAAGCUAUGCAGCCCUAGCUUUAAUCCAUUUGCAGAGCUGCCAACGUGGACAAAGUGGCGUU-UGCCAAACAGUUGGCAUCGCCACCA (((((.....)))))....(((((.......(((((.((.((............)))).)))))....)))))....((((((..-((((((....)))))).)))))).. ( -34.10) >consensus UAGAAGCGGAUCUUAAAAAUCCGCAAGCGAUGCAGCCCUAGCUUAAGUCCAUUUCCAGAGCUGCCAACGUGGGCAAAGUGGCCCUAUGCCAACCAGUUGGCAUCGCCACCA .....((((((.......))))))..(((((((.((((((((((.............)))))).......))))....((((.....))))........)))))))..... (-28.67 = -29.27 + 0.60)

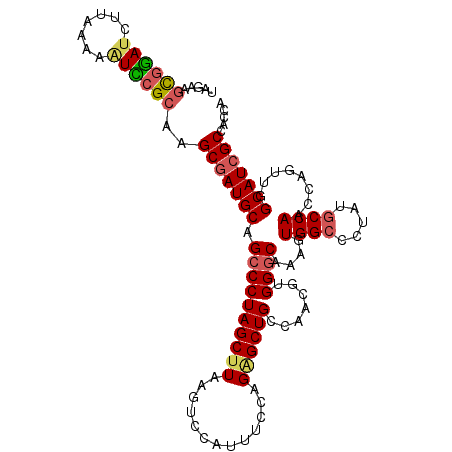
| Location | 1,297,141 – 1,297,252 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 87.65 |
| Mean single sequence MFE | -42.23 |
| Consensus MFE | -33.44 |
| Energy contribution | -34.72 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.79 |
| SVM decision value | 2.19 |
| SVM RNA-class probability | 0.989997 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1297141 111 - 23771897 UGGUGGCGAUGCCAACUGGUUGGGAUAGGGCCACAUUGCCCACGUUGGCAGCCCUGACAAUGGGCUUAAGCUAGGGCUGCAUCGCUUGCGGACUUUUAAGAUCCGCUUUGA ....((((((.((((....))))....((((......))))......((((((((.......(((....))))))))))))))))).(((((((....)).)))))..... ( -43.01) >DroSec_CAF1 926 111 - 1 UGGUGGCGAUGCCAACUGGUUGGCAUAGGGCCACUUUGCCCACGUUGGCAGCUCUGGAAAUUAACUUAAGCUAGGGCUGCAUCGCUUUCGGAUUUUUAAGAUCCGCUCCUA .((.(((((((((((....)))))...((((......))))......((((((((((...(((...))).)))))))))))))))...((((((.....))))))).)).. ( -44.60) >DroSim_CAF1 4227 111 - 1 UGGUGGCGAUGCCAACUGGUUGGCAUAGGGCCACUUUGCCCACGUUGGCAGCUCUGGAAAUGAACUUAAGCUAGGGCUGCAUCGCUUGCGGAUUUUUAAGAUCCGCUUCUA ....(((((((((((....)))))...((((......))))......((((((((((.............)))))))))))))))).(((((((.....)))))))..... ( -47.42) >DroEre_CAF1 902 110 - 1 UGGUGGCGAUGCCAACUGAUUGGCA-AGAGCCACUUUGCCCACGUUGGCAGCGCUGUAAAUGGAUUCAAGCUAGGGCUGCAUCGCUUGCGAAUUUUAAAGCUCCACUUCUA .(((((.(.(((((((((...((((-((......)))))))).))))))).)(((......((((((.(((....)))(((.....)))))))))...))).))))).... ( -34.90) >DroYak_CAF1 4140 110 - 1 UGGUGGCGAUGCCAACUGUUUGGCA-AACGCCACUUUGUCCACGUUGGCAGCUCUGCAAAUGGAUUAAAGCUAGGGCUGCAUAGCUUGCGAAUUUUUAAGAUUUGCUCUUA .(((((((.((((((....))))))-..)))))))........((((((((((((((............)).)))))))).))))..(((((((.....)))))))..... ( -41.20) >consensus UGGUGGCGAUGCCAACUGGUUGGCAUAGGGCCACUUUGCCCACGUUGGCAGCUCUGGAAAUGGACUUAAGCUAGGGCUGCAUCGCUUGCGGAUUUUUAAGAUCCGCUUCUA ..(((((.(((((((....)))))))...))))).........((..(((((((((...............)))))))))...))..(((((((.....)))))))..... (-33.44 = -34.72 + 1.28)

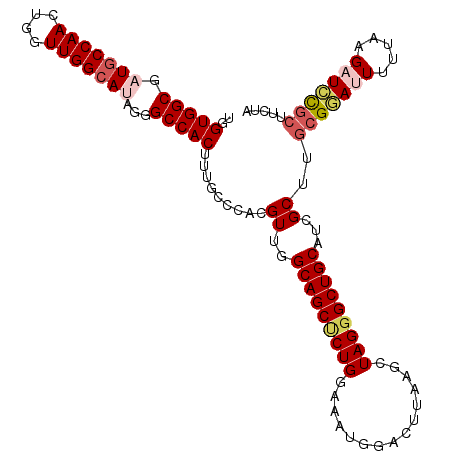
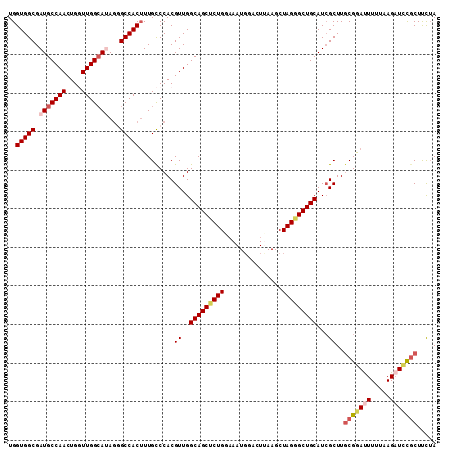
| Location | 1,297,181 – 1,297,277 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 90.93 |
| Mean single sequence MFE | -31.05 |
| Consensus MFE | -25.10 |
| Energy contribution | -25.98 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958338 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1297181 96 + 23771897 GCUUAAGCCCAUUGUCAGGGCUGCCAACGUGGGCAAUGUGGCCCUAUCCCAACCAGUUGGCAUCGCCACCAUUAUUUAUUGGUUUUUUGUUGCUGG ((...(((((.......)))))((((((..((((......))))...........))))))...)).((((........))))............. ( -27.92) >DroSec_CAF1 966 96 + 1 GCUUAAGUUAAUUUCCAGAGCUGCCAACGUGGGCAAAGUGGCCCUAUGCCAACCAGUUGGCAUCGCCACCAUUAUUUGUUGGUUUUUUGUUGCUGG ..............(((((((.(((((((........(((((...(((((((....))))))).))))).......))))))).....))).)))) ( -31.36) >DroSim_CAF1 4267 96 + 1 GCUUAAGUUCAUUUCCAGAGCUGCCAACGUGGGCAAAGUGGCCCUAUGCCAACCAGUUGGCAUCGCCACCAUUAUUUGUUGGUUUUUUGUUGCUGG ..............(((((((.(((((((........(((((...(((((((....))))))).))))).......))))))).....))).)))) ( -31.36) >DroEre_CAF1 942 95 + 1 GCUUGAAUCCAUUUACAGCGCUGCCAACGUGGGCAAAGUGGCUCU-UGCCAAUCAGUUGGCAUCGCCACCAUUAUUUGUUGGUUUUUUGUUGCUGG ...............((((((.(((((((........(((((...-((((((....))))))..))))).......))))))).....).))))). ( -30.66) >DroYak_CAF1 4180 95 + 1 GCUUUAAUCCAUUUGCAGAGCUGCCAACGUGGACAAAGUGGCGUU-UGCCAAACAGUUGGCAUCGCCACCAUUAUUUGUUGGUUUUUUGUUGCUGG ........(((...((((((..(((((((........((((((..-((((((....)))))).)))))).......))))))).))))))...))) ( -33.96) >consensus GCUUAAGUCCAUUUCCAGAGCUGCCAACGUGGGCAAAGUGGCCCUAUGCCAACCAGUUGGCAUCGCCACCAUUAUUUGUUGGUUUUUUGUUGCUGG ...............((((((.(((((((........(((((...(((((((....))))))).))))).......))))))).....))).))). (-25.10 = -25.98 + 0.88)


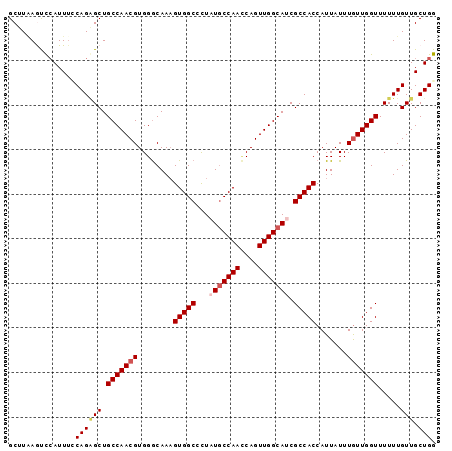
| Location | 1,297,181 – 1,297,277 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 90.93 |
| Mean single sequence MFE | -29.20 |
| Consensus MFE | -22.26 |
| Energy contribution | -22.70 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.957518 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1297181 96 - 23771897 CCAGCAACAAAAAACCAAUAAAUAAUGGUGGCGAUGCCAACUGGUUGGGAUAGGGCCACAUUGCCCACGUUGGCAGCCCUGACAAUGGGCUUAAGC (((((.........(((((......((((......))))....)))))....((((......))))..))))).(((((.......)))))..... ( -30.60) >DroSec_CAF1 966 96 - 1 CCAGCAACAAAAAACCAACAAAUAAUGGUGGCGAUGCCAACUGGUUGGCAUAGGGCCACUUUGCCCACGUUGGCAGCUCUGGAAAUUAACUUAAGC ((((......................((((((.(((((((....)))))))...))))))(((((......)))))..)))).............. ( -28.90) >DroSim_CAF1 4267 96 - 1 CCAGCAACAAAAAACCAACAAAUAAUGGUGGCGAUGCCAACUGGUUGGCAUAGGGCCACUUUGCCCACGUUGGCAGCUCUGGAAAUGAACUUAAGC ((((......................((((((.(((((((....)))))))...))))))(((((......)))))..)))).............. ( -28.90) >DroEre_CAF1 942 95 - 1 CCAGCAACAAAAAACCAACAAAUAAUGGUGGCGAUGCCAACUGAUUGGCA-AGAGCCACUUUGCCCACGUUGGCAGCGCUGUAAAUGGAUUCAAGC (((...((.....((((........))))((((.(((((((((...((((-((......)))))))).))))))).))))))...)))........ ( -30.10) >DroYak_CAF1 4180 95 - 1 CCAGCAACAAAAAACCAACAAAUAAUGGUGGCGAUGCCAACUGUUUGGCA-AACGCCACUUUGUCCACGUUGGCAGCUCUGCAAAUGGAUUAAAGC ((((((........(((((.......(((((((.((((((....))))))-..)))))))........)))))......)))...)))........ ( -27.50) >consensus CCAGCAACAAAAAACCAACAAAUAAUGGUGGCGAUGCCAACUGGUUGGCAUAGGGCCACUUUGCCCACGUUGGCAGCUCUGGAAAUGGACUUAAGC ...((......................(((((.(((((((....)))))))...)))))..((((......))))))................... (-22.26 = -22.70 + 0.44)

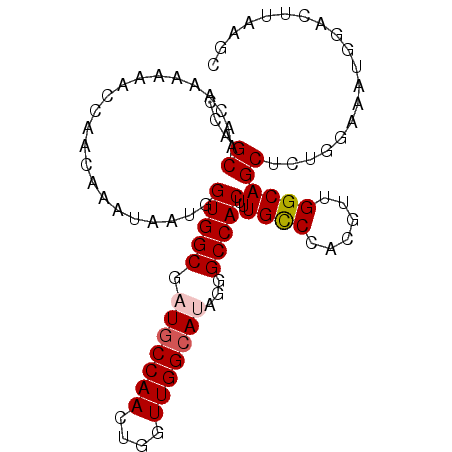
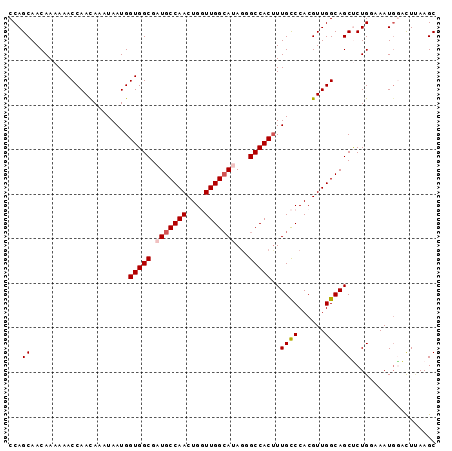
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:42:03 2006