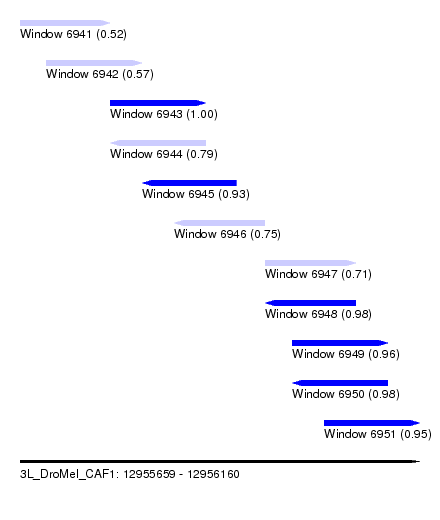
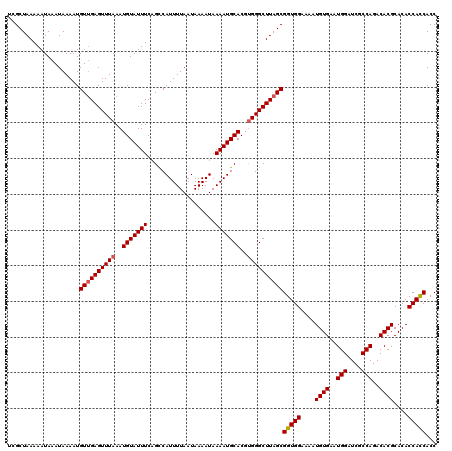
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,955,659 – 12,956,160 |
| Length | 501 |
| Max. P | 0.998833 |

| Location | 12,955,659 – 12,955,772 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.74 |
| Mean single sequence MFE | -16.94 |
| Consensus MFE | -12.28 |
| Energy contribution | -12.64 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.72 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.524690 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12955659 113 + 23771897 U-----AUUUGCUUAACACAA--ACAUCAAGAAGAUAACAAAAUAUUCCAACACGAAAAUAUGCAGAAAUCACAGUGAAAAUGGGAGAAACUGUGAACACUGUAGAACUACUGGUUAAAG .-----......(((((.((.--..(((.....))).........................(((((...(((((((.............)))))))...))))).......))))))).. ( -16.32) >DroSec_CAF1 72857 115 + 1 U-----AUUUGCUUAACACAAAAACAGCAAGAAGAUAACAAAAUAUUCCAACACAAAAAUAUGCAGAAAUCACAGUGAAAAUGGGAGGAACUGUGAAAACUGUAAAAAUACUGGUUAAAG .-----.((((((............))))))..................(((.........(((((...(((((((.............)))))))...))))).........))).... ( -16.69) >DroSim_CAF1 77693 99 + 1 U-----AUUUGCUUAACACAAAAACAGCAAGAAGAUAACAAA----------------AUAUGCAGAAAUCACAGUGAAAAUGGUAGGAACUGUGAAAACUGUAAAAAUACUGGUUAAAG .-----.((((((............))))))....((((...----------------...(((((...(((((((.............)))))))...))))).........))))... ( -16.78) >DroEre_CAF1 79063 109 + 1 U-----AUUUGCUUAACACAAACACAGCAAGAAGAUAAUAAAAUAUGCCAACAUGAAAAUAUGCAGAAAUCACAGUUUAAAUGGGAGGAAAUGUGAAAACUGUGAAAAUACUG------G .-----.((((((............))))))............((((....))))........(((...(((((((((..(((........)))..))))))))).....)))------. ( -14.70) >DroYak_CAF1 76854 120 + 1 UAUGGUAUUUGCUUAACACAAGCACAGCAAGAAGAUAAUAAAAUAUGCCAACAUGAAAAUAUGCAGAAAUCACAGUUAAAAUGGGAGAAAAUGUGAAAACUGUGAAAAUACCAGUGGAAG ..((((((((((((.....))))......................(((....((....))..)))....((((((((...(((........)))...))))))))))))))))....... ( -20.20) >consensus U_____AUUUGCUUAACACAAAAACAGCAAGAAGAUAACAAAAUAUGCCAACACGAAAAUAUGCAGAAAUCACAGUGAAAAUGGGAGGAACUGUGAAAACUGUAAAAAUACUGGUUAAAG .......((((((............))))))..............................(((((...(((((((.............)))))))...)))))................ (-12.28 = -12.64 + 0.36)

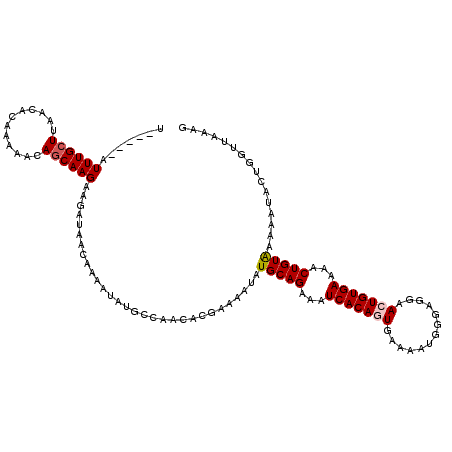

| Location | 12,955,692 – 12,955,812 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.58 |
| Mean single sequence MFE | -18.77 |
| Consensus MFE | -14.37 |
| Energy contribution | -15.65 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.571337 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12955692 120 + 23771897 AAAUAUUCCAACACGAAAAUAUGCAGAAAUCACAGUGAAAAUGGGAGAAACUGUGAACACUGUAGAACUACUGGUUAAAGUCGCUAAAAAUAAAUAAAAUGUCGAGUUUAAAUGUAUUUC .....................(((((...(((((((.............)))))))...)))))(((.(((...(((((.((((................).))).)))))..))).))) ( -21.21) >DroSec_CAF1 72892 120 + 1 AAAUAUUCCAACACAAAAAUAUGCAGAAAUCACAGUGAAAAUGGGAGGAACUGUGAAAACUGUAAAAAUACUGGUUAAAGUCGCUAAAAAUAAAUAAAAUGUUGAGUUUAAAUGUAUUUC .....................(((((...(((((((.............)))))))...))))).((((((...(((((.((((................).))).)))))..)))))). ( -20.31) >DroSim_CAF1 77728 104 + 1 AA----------------AUAUGCAGAAAUCACAGUGAAAAUGGUAGGAACUGUGAAAACUGUAAAAAUACUGGUUAAAGUCGCUAAAAAUAAAUAAAAUGUUGAGUUUAAAUGUAUUUC ..----------------...(((((...(((((((.............)))))))...))))).((((((...(((((.((((................).))).)))))..)))))). ( -20.31) >DroEre_CAF1 79098 114 + 1 AAAUAUGCCAACAUGAAAAUAUGCAGAAAUCACAGUUUAAAUGGGAGGAAAUGUGAAAACUGUGAAAAUACUG------GUCGCUAAAAAUAAAUAAAAUGUUGAGUUUAAAUGUAUUUC ...((.((((((((....))...(((...(((((((((..(((........)))..))))))))).....)))------.....................)))).)).)).......... ( -14.70) >DroYak_CAF1 76894 120 + 1 AAAUAUGCCAACAUGAAAAUAUGCAGAAAUCACAGUUAAAAUGGGAGAAAAUGUGAAAACUGUGAAAAUACCAGUGGAAGUCGCUAAAAAUAAAUAAAAUGUUGAGUUUAAAUGUAUUUC ...((.((((((((...............((((((((...(((........)))...)))))))).......((((.....)))).............)))))).)).)).......... ( -17.30) >consensus AAAUAUGCCAACACGAAAAUAUGCAGAAAUCACAGUGAAAAUGGGAGGAACUGUGAAAACUGUAAAAAUACUGGUUAAAGUCGCUAAAAAUAAAUAAAAUGUUGAGUUUAAAUGUAUUUC .....................(((((...(((((((.............)))))))...))))).((((((...(((((.((((................).))).)))))..)))))). (-14.37 = -15.65 + 1.28)


| Location | 12,955,772 – 12,955,892 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.17 |
| Mean single sequence MFE | -28.13 |
| Consensus MFE | -28.01 |
| Energy contribution | -28.25 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.09 |
| Structure conservation index | 1.00 |
| SVM decision value | 3.24 |
| SVM RNA-class probability | 0.998833 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12955772 120 + 23771897 UCGCUAAAAAUAAAUAAAAUGUCGAGUUUAAAUGUAUUUCAGCCAUUUUAAUAAAAUAAAAUGCACGUGGGCUUAGCGGUGGAAAAUGUGAAUGGAUCGCCAGACACGCACACCACCACC ....................((..((((((..(((((((...................)))))))..))))))..))(((((....((((..(((....)))..))))....)))))... ( -27.31) >DroSec_CAF1 72972 120 + 1 UCGCUAAAAAUAAAUAAAAUGUUGAGUUUAAAUGUAUUUCAGCCAUUUUAAUAAAAUAAAAUGCACGUGGGCUUAGCGGUGGAAAAUGUGAAUGGAUCGCCAGACACGCACACCACCACC ....................((((((((((..(((((((...................)))))))..))))))))))(((((....((((..(((....)))..))))....)))))... ( -29.71) >DroSim_CAF1 77792 120 + 1 UCGCUAAAAAUAAAUAAAAUGUUGAGUUUAAAUGUAUUUCAGCCAUUUUAAUAAAAUAAAAUGCACGUGGGCUUAGCGGUGGAAAAUGUGAAUGGAUCGCCAGACACGCACACCACCACC ....................((((((((((..(((((((...................)))))))..))))))))))(((((....((((..(((....)))..))))....)))))... ( -29.71) >DroEre_CAF1 79172 120 + 1 UCGCUAAAAAUAAAUAAAAUGUUGAGUUUAAAUGUAUUUCUGCCAUUUUAAUAAAAUAAAAUGCACGUGGGCUUAGCGGUGGAAAAUGUGAAUGGAUCGCCAGACACGCACACCACCACC ....................((((((((((..(((((((...................)))))))..))))))))))(((((....((((..(((....)))..))))....)))))... ( -29.71) >DroYak_CAF1 76974 120 + 1 UCGCUAAAAAUAAAUAAAAUGUUGAGUUUAAAUGUAUUUCUGUCAUUUUAAUAAAAUAAAAUGCACGCGGGCUUAGCGGUGGAAAAUGUGAAUGGAUCGCCAGACACGCACACCAUCACC ..(((((...........(((((.......)))))...((((((((((((......)))))))...))))).)))))(((((....((((..(((....)))..))))....)))))... ( -24.20) >consensus UCGCUAAAAAUAAAUAAAAUGUUGAGUUUAAAUGUAUUUCAGCCAUUUUAAUAAAAUAAAAUGCACGUGGGCUUAGCGGUGGAAAAUGUGAAUGGAUCGCCAGACACGCACACCACCACC ....................((((((((((..(((((((...................)))))))..))))))))))(((((....((((..(((....)))..))))....)))))... (-28.01 = -28.25 + 0.24)

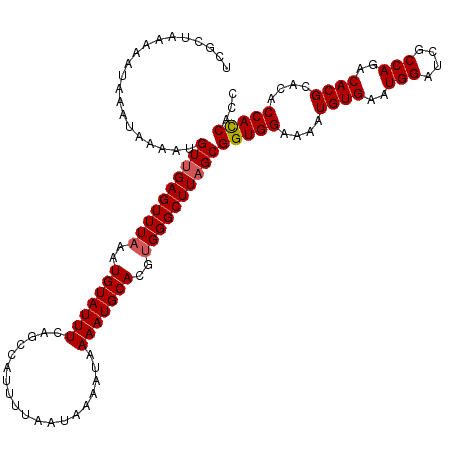
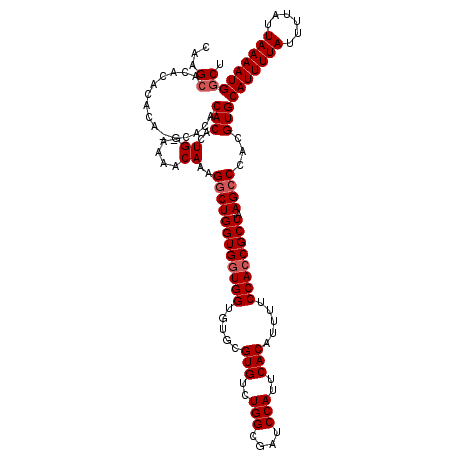
| Location | 12,955,772 – 12,955,892 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.17 |
| Mean single sequence MFE | -27.72 |
| Consensus MFE | -25.10 |
| Energy contribution | -25.90 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.791680 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12955772 120 - 23771897 GGUGGUGGUGUGCGUGUCUGGCGAUCCAUUCACAUUUUCCACCGCUAAGCCCACGUGCAUUUUAUUUUAUUAAAAUGGCUGAAAUACAUUUAAACUCGACAUUUUAUUUAUUUUUAGCGA ((((((((.....(((..(((....)))..))).....))))))))...........(((((((......)))))))(((((((......((((........)))).....))))))).. ( -29.40) >DroSec_CAF1 72972 120 - 1 GGUGGUGGUGUGCGUGUCUGGCGAUCCAUUCACAUUUUCCACCGCUAAGCCCACGUGCAUUUUAUUUUAUUAAAAUGGCUGAAAUACAUUUAAACUCAACAUUUUAUUUAUUUUUAGCGA ((((((((.....(((..(((....)))..))).....))))))))...........(((((((......)))))))(((((((......((((........)))).....))))))).. ( -29.40) >DroSim_CAF1 77792 120 - 1 GGUGGUGGUGUGCGUGUCUGGCGAUCCAUUCACAUUUUCCACCGCUAAGCCCACGUGCAUUUUAUUUUAUUAAAAUGGCUGAAAUACAUUUAAACUCAACAUUUUAUUUAUUUUUAGCGA ((((((((.....(((..(((....)))..))).....))))))))...........(((((((......)))))))(((((((......((((........)))).....))))))).. ( -29.40) >DroEre_CAF1 79172 120 - 1 GGUGGUGGUGUGCGUGUCUGGCGAUCCAUUCACAUUUUCCACCGCUAAGCCCACGUGCAUUUUAUUUUAUUAAAAUGGCAGAAAUACAUUUAAACUCAACAUUUUAUUUAUUUUUAGCGA ((((((((.....(((..(((....)))..))).....))))))))...........(((((((......)))))))(((((((((....((((........))))..))))))).)).. ( -27.00) >DroYak_CAF1 76974 120 - 1 GGUGAUGGUGUGCGUGUCUGGCGAUCCAUUCACAUUUUCCACCGCUAAGCCCGCGUGCAUUUUAUUUUAUUAAAAUGACAGAAAUACAUUUAAACUCAACAUUUUAUUUAUUUUUAGCGA ((((..((((((.(((((....))..))).))))))...))))........(((((.(((((((......)))))))))(((((((....((((........))))..))))))).))). ( -23.40) >consensus GGUGGUGGUGUGCGUGUCUGGCGAUCCAUUCACAUUUUCCACCGCUAAGCCCACGUGCAUUUUAUUUUAUUAAAAUGGCUGAAAUACAUUUAAACUCAACAUUUUAUUUAUUUUUAGCGA ((((((((.....(((..(((....)))..))).....))))))))...........(((((((......)))))))(((((((......((((........)))).....))))))).. (-25.10 = -25.90 + 0.80)

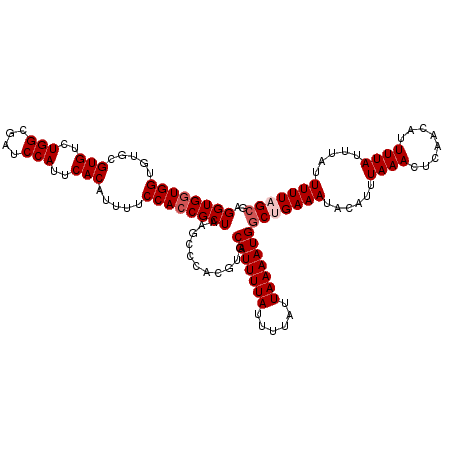
| Location | 12,955,812 – 12,955,930 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.00 |
| Mean single sequence MFE | -34.58 |
| Consensus MFE | -28.98 |
| Energy contribution | -29.72 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.930451 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12955812 118 - 23771897 CAAGCACACACACA--CACACACACUGGAAAACAAAGGCUGGUGGUGGUGUGCGUGUCUGGCGAUCCAUUCACAUUUUCCACCGCUAAGCCCACGUGCAUUUUAUUUUAUUAAAAUGGCU ..(((.........--....(((..((.....))..((((((((((((.....(((..(((....)))..))).....)))))))).))))...)))(((((((......)))))))))) ( -34.60) >DroSec_CAF1 73012 118 - 1 CAAGCACACGCACA--CACACACACUGGAAAACAAAGGCUGGUGGUGGUGUGCGUGUCUGGCGAUCCAUUCACAUUUUCCACCGCUAAGCCCACGUGCAUUUUAUUUUAUUAAAAUGGCU ..((((((((((((--(.(((.(((..(..........)..)))))))))))))))).(((((...................)))))..........(((((((......)))))))))) ( -37.41) >DroSim_CAF1 77832 120 - 1 CAAGCACACACACAACCACACACACUGGAAAACAAAGGCUGGUGGUGGUGUGCGUGUCUGGCGAUCCAUUCACAUUUUCCACCGCUAAGCCCACGUGCAUUUUAUUUUAUUAAAAUGGCU ..(((.....(((..(((.......)))........((((((((((((.....(((..(((....)))..))).....)))))))).))))...)))(((((((......)))))))))) ( -36.60) >DroYak_CAF1 77014 112 - 1 CAAGCA------CA--UGCACACACUGGGAAACAAAGACUGGUGAUGGUGUGCGUGUCUGGCGAUCCAUUCACAUUUUCCACCGCUAAGCCCGCGUGCAUUUUAUUUUAUUAAAAUGACA ...(((------(.--.((((.(((..(..........)..)))...))))(((.(..(((((...................)))))..).)))))))((((((......)))))).... ( -29.71) >consensus CAAGCACACACACA__CACACACACUGGAAAACAAAGGCUGGUGGUGGUGUGCGUGUCUGGCGAUCCAUUCACAUUUUCCACCGCUAAGCCCACGUGCAUUUUAUUUUAUUAAAAUGGCU ...((...............(((..((.....))..((((((((((((.....(((..(((....)))..))).....)))))))).))))...)))(((((((......))))))))). (-28.98 = -29.72 + 0.75)

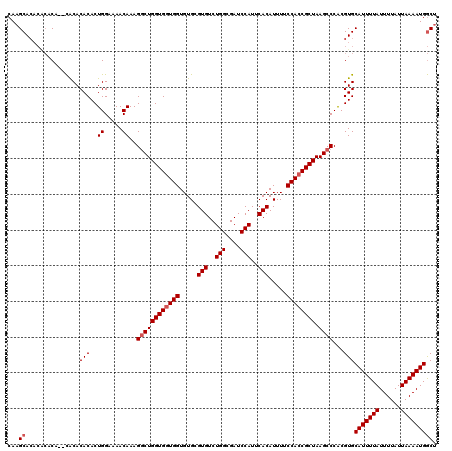
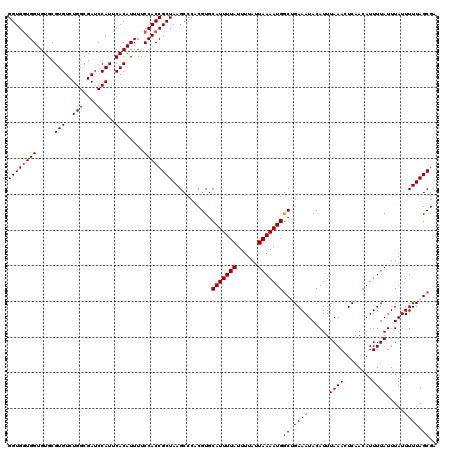
| Location | 12,955,852 – 12,955,966 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 92.32 |
| Mean single sequence MFE | -33.83 |
| Consensus MFE | -25.65 |
| Energy contribution | -26.90 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.753730 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12955852 114 - 23771897 GAAGCACACAUACCUUUUCAGGUAAAGGGUGAAAUUCAAGCACACACACA--CACACACACUGGAAAACAAAGGCUGGUGGUGGUGUGCGUGUCUGGCGAUCCAUUCACAUUUUCC (((.(((...(((((....)))))....)))...)))..((((((.(((.--(((.((..((.........))..))))))))))))))(((..(((....)))..)))....... ( -34.40) >DroSec_CAF1 73052 114 - 1 GAAACACACAUACCUUUUCAGGUAAAGGGAGAAAUUCAAGCACACGCACA--CACACACACUGGAAAACAAAGGCUGGUGGUGGUGUGCGUGUCUGGCGAUCCAUUCACAUUUUCC ..........(((((....)))))...((((((......(((((((((((--(.(((.(((..(..........)..))))))))))))))))...))((.....))...)))))) ( -37.50) >DroSim_CAF1 77872 116 - 1 GAAACACACAUACCUUUUCAGGUAAAGGGAGAAAUUCAAGCACACACACAACCACACACACUGGAAAACAAAGGCUGGUGGUGGUGUGCGUGUCUGGCGAUCCAUUCACAUUUUCC (((.......(((((....)))))...(((.....(((..(((.(((((.(((((.((..((.........))..))))))).))))).)))..)))...))).)))......... ( -33.50) >DroYak_CAF1 77054 108 - 1 CAAACACGCAUACCUUUUCAGGUAAAGGGAGAAAUUCAAGCA------CA--UGCACACACUGGGAAACAAAGACUGGUGAUGGUGUGCGUGUCUGGCGAUCCAUUCACAUUUUCC ((.(((((((((((.......((....(((....)))..((.------..--.)))).(((..(..........)..)))..))))))))))).)).................... ( -29.90) >consensus GAAACACACAUACCUUUUCAGGUAAAGGGAGAAAUUCAAGCACACACACA__CACACACACUGGAAAACAAAGGCUGGUGGUGGUGUGCGUGUCUGGCGAUCCAUUCACAUUUUCC ..........(((((....)))))...((((((......((((((.........(((.(((..(..........)..))))))))))))(((..(((....)))..))).)))))) (-25.65 = -26.90 + 1.25)

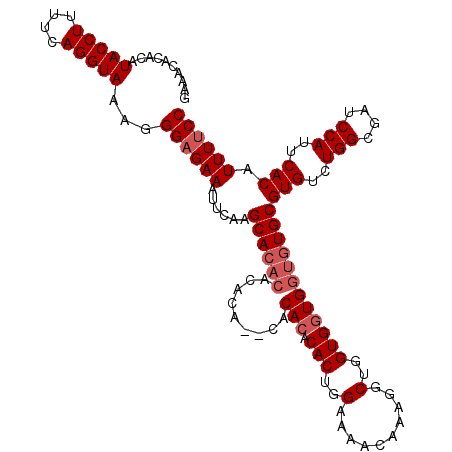
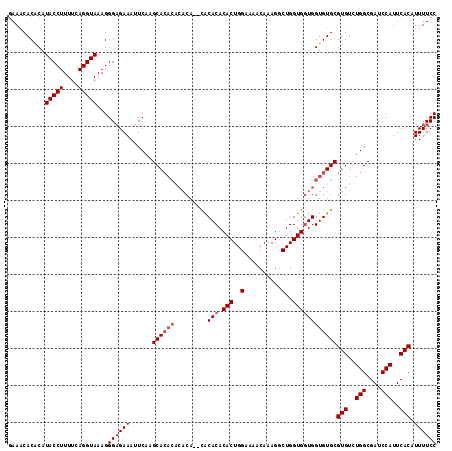
| Location | 12,955,966 – 12,956,080 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.72 |
| Mean single sequence MFE | -33.58 |
| Consensus MFE | -31.08 |
| Energy contribution | -30.95 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.713567 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12955966 114 + 23771897 U----GUGGGU--GUGGGAGUGCUGAGUGUGUUUAAAUGCAAAUUACAUUUCAUUUCAUUACUGCGCGCGGUAUUUUCACGCUGGGCUUGAAAAUACGGGAAUGCGCAGCGGGAAAAUAA .----......--...((((((.(((...(((......)))..))))))))).((((.(..((((((((.(((((((((.((...)).))))))))).)...)))))))..))))).... ( -32.70) >DroSec_CAF1 73166 113 + 1 C----GUGGGUUUGUGGGAGUGCUGAGUGUGUUUAAAUGCAAAUUACAUUUCAUUUCAUUACUGCGCGCGGUAUUUUCACGCUGGGCUUGAAAAUACGGGAAUGCGCAGCGGGAAA---A .----((((.((((((.(((..(.....)..)))...))))))))))......((((.(..((((((((.(((((((((.((...)).))))))))).)...)))))))..)))))---. ( -34.20) >DroSim_CAF1 77988 114 + 1 U----GUGGGUGUGUGGGAGUGCUGAGUGUGUUUAAAUGCAAAUUACAUUUCAUUUCAUUACUGCGCGCGGUAUUUUCACGCUGGGCUUGAAAAUACGGGAAUGCGCAGCGGGAAAA--A (----(((..(((((..(((..(.....)..)))..)))))...)))).....((((.(..((((((((.(((((((((.((...)).))))))))).)...)))))))..))))).--. ( -34.50) >DroYak_CAF1 77162 117 + 1 UGAAUGUGUGUGUGGGGGAGUGCUGAGUGUGUUUAAAUGCAAAUUACAUUUCAUUUCAUUACUGUGCGCGGUAUUUUCACGCUGGGCUUGGAAAUACAAGAAUGCGCAGCGGGAAA---A .(((((((..((((...(((..(.....)..)))...))))...)))))))..((((.(..(((((((..(((((((((.((...)).))))))))).....)))))))..)))))---. ( -32.90) >consensus U____GUGGGUGUGUGGGAGUGCUGAGUGUGUUUAAAUGCAAAUUACAUUUCAUUUCAUUACUGCGCGCGGUAUUUUCACGCUGGGCUUGAAAAUACGGGAAUGCGCAGCGGGAAA___A ................((((((.(((...(((......)))..))))))))).((((.(..((((((((.(((((((((.((...)).))))))))).)...)))))))..))))).... (-31.08 = -30.95 + -0.13)

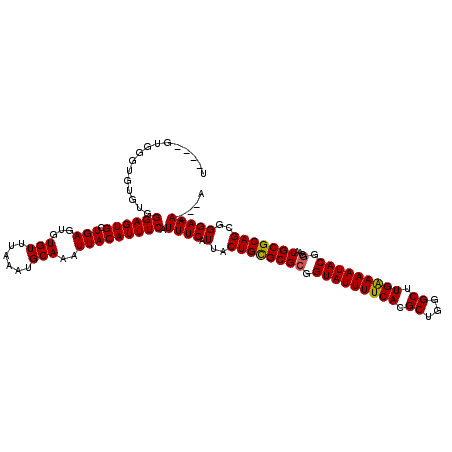
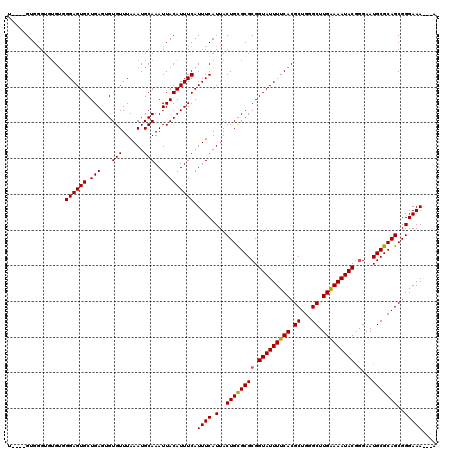
| Location | 12,955,966 – 12,956,080 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.72 |
| Mean single sequence MFE | -23.72 |
| Consensus MFE | -22.06 |
| Energy contribution | -22.38 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.980933 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12955966 114 - 23771897 UUAUUUUCCCGCUGCGCAUUCCCGUAUUUUCAAGCCCAGCGUGAAAAUACCGCGCGCAGUAAUGAAAUGAAAUGUAAUUUGCAUUUAAACACACUCAGCACUCCCAC--ACCCAC----A .((((((...(((((((....(.(((((((((.((...)).))))))))).).)))))))...))))))((((((.....)))))).....................--......----. ( -25.90) >DroSec_CAF1 73166 113 - 1 U---UUUCCCGCUGCGCAUUCCCGUAUUUUCAAGCCCAGCGUGAAAAUACCGCGCGCAGUAAUGAAAUGAAAUGUAAUUUGCAUUUAAACACACUCAGCACUCCCACAAACCCAC----G .---((((..(((((((....(.(((((((((.((...)).))))))))).).)))))))...))))..((((((.....)))))).............................----. ( -25.10) >DroSim_CAF1 77988 114 - 1 U--UUUUCCCGCUGCGCAUUCCCGUAUUUUCAAGCCCAGCGUGAAAAUACCGCGCGCAGUAAUGAAAUGAAAUGUAAUUUGCAUUUAAACACACUCAGCACUCCCACACACCCAC----A .--.((((..(((((((....(.(((((((((.((...)).))))))))).).)))))))...))))..((((((.....)))))).............................----. ( -25.10) >DroYak_CAF1 77162 117 - 1 U---UUUCCCGCUGCGCAUUCUUGUAUUUCCAAGCCCAGCGUGAAAAUACCGCGCACAGUAAUGAAAUGAAAUGUAAUUUGCAUUUAAACACACUCAGCACUCCCCCACACACACAUUCA .---......((((.(........((((((...((...(((((.......)))))...))...))))))((((((.....)))))).......).))))..................... ( -18.80) >consensus U___UUUCCCGCUGCGCAUUCCCGUAUUUUCAAGCCCAGCGUGAAAAUACCGCGCGCAGUAAUGAAAUGAAAUGUAAUUUGCAUUUAAACACACUCAGCACUCCCACACACCCAC____A ....((((..(((((((....(.(((((((((.((...)).))))))))).).)))))))...))))..((((((.....)))))).................................. (-22.06 = -22.38 + 0.31)

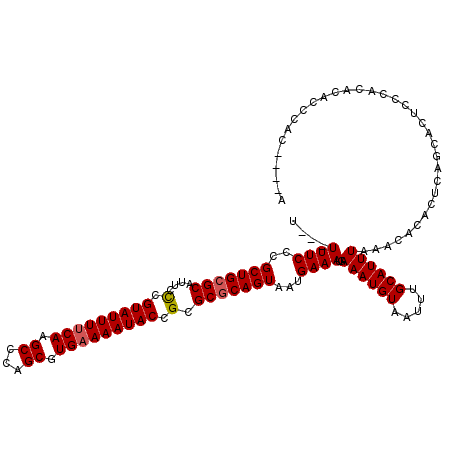
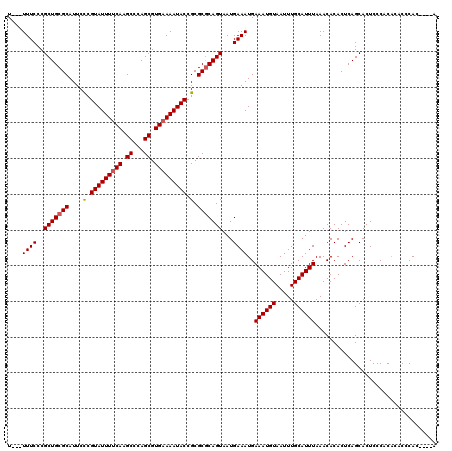
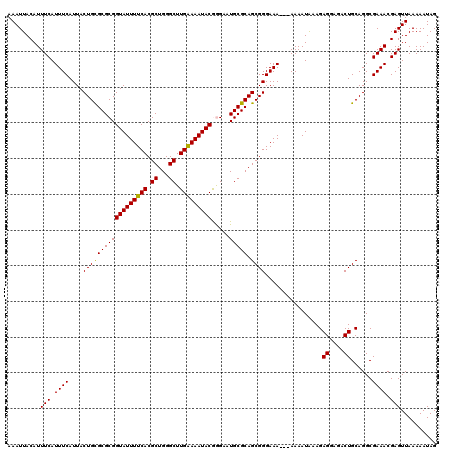
| Location | 12,956,000 – 12,956,120 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.51 |
| Mean single sequence MFE | -28.53 |
| Consensus MFE | -25.98 |
| Energy contribution | -25.50 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962274 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12956000 120 + 23771897 AAAUUACAUUUCAUUUCAUUACUGCGCGCGGUAUUUUCACGCUGGGCUUGAAAAUACGGGAAUGCGCAGCGGGAAAAUAAAAAUAAAGAGGAGACUGCAGGCGAAACGAGUUAAAAAUAG .((((.(.((((.((((.(..((((((((.(((((((((.((...)).))))))))).)...)))))))..)))))............((....))......)))).)))))........ ( -29.90) >DroSec_CAF1 73202 117 + 1 AAAUUACAUUUCAUUUCAUUACUGCGCGCGGUAUUUUCACGCUGGGCUUGAAAAUACGGGAAUGCGCAGCGGGAAA---AAAAUAAAGAGGAGACUGCAGGCGAAACGAGUUAAAAAUAG .((((.(.((((.((((.(..((((((((.(((((((((.((...)).))))))))).)...)))))))..)))))---.........((....))......)))).)))))........ ( -29.90) >DroSim_CAF1 78024 118 + 1 AAAUUACAUUUCAUUUCAUUACUGCGCGCGGUAUUUUCACGCUGGGCUUGAAAAUACGGGAAUGCGCAGCGGGAAAA--AAAAUGAAGAGGAGACUGCAGGCGAAACGAGUUAAAAAUAG ......(((((..((((.(..((((((((.(((((((((.((...)).))))))))).)...)))))))..))))).--.)))))...((....))........................ ( -31.10) >DroEre_CAF1 79402 115 + 1 AAAUUACAUUUCAUUUCAUUACUGUGCGCGGUAUUUUCACGCUGGGCUUGGAAAUACAAGAAUGCGCAGCGGGAAA---AAAAUAAAGAGA--CCUGCAGGCGAAACGAGUUAAAAAUAG .........(((.((((.......(((((((((((((((.((...)).))))))))).....))))))(((((...---............--)))))....)))).))).......... ( -26.66) >DroYak_CAF1 77202 115 + 1 AAAUUACAUUUCAUUUCAUUACUGUGCGCGGUAUUUUCACGCUGGGCUUGGAAAUACAAGAAUGCGCAGCGGGAAA---AAAAUAAAGAGC--ACUGCAGGCGAAACGAGUUAAAAAUAG .........(((.((((.((.(.((((.(..(((((((.(((((.(((((......)))....)).))))).))..---.)))))..).))--)).).))..)))).))).......... ( -25.10) >consensus AAAUUACAUUUCAUUUCAUUACUGCGCGCGGUAUUUUCACGCUGGGCUUGAAAAUACGGGAAUGCGCAGCGGGAAA___AAAAUAAAGAGGAGACUGCAGGCGAAACGAGUUAAAAAUAG .........(((.((((....((((((((((((((((((.((...)).))))))))).....))))).))))................((....))......)))).))).......... (-25.98 = -25.50 + -0.48)

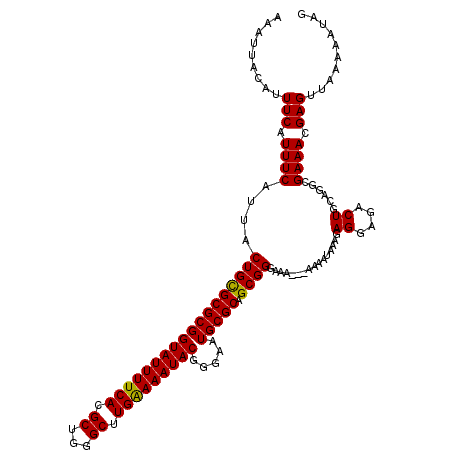
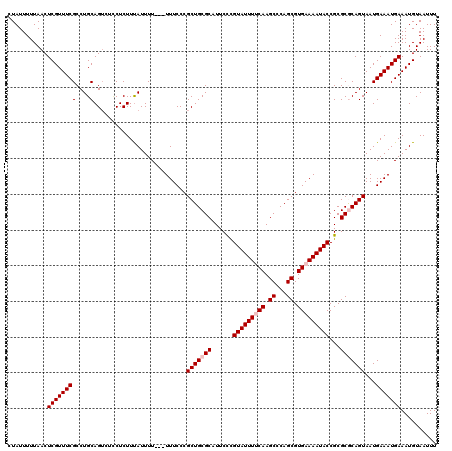
| Location | 12,956,000 – 12,956,120 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.51 |
| Mean single sequence MFE | -25.90 |
| Consensus MFE | -22.16 |
| Energy contribution | -22.96 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.981892 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12956000 120 - 23771897 CUAUUUUUAACUCGUUUCGCCUGCAGUCUCCUCUUUAUUUUUAUUUUCCCGCUGCGCAUUCCCGUAUUUUCAAGCCCAGCGUGAAAAUACCGCGCGCAGUAAUGAAAUGAAAUGUAAUUU ...........((((((((....)..........................(((((((....(.(((((((((.((...)).))))))))).).)))))))...))))))).......... ( -26.80) >DroSec_CAF1 73202 117 - 1 CUAUUUUUAACUCGUUUCGCCUGCAGUCUCCUCUUUAUUUU---UUUCCCGCUGCGCAUUCCCGUAUUUUCAAGCCCAGCGUGAAAAUACCGCGCGCAGUAAUGAAAUGAAAUGUAAUUU ...........((((((((....).................---......(((((((....(.(((((((((.((...)).))))))))).).)))))))...))))))).......... ( -26.80) >DroSim_CAF1 78024 118 - 1 CUAUUUUUAACUCGUUUCGCCUGCAGUCUCCUCUUCAUUUU--UUUUCCCGCUGCGCAUUCCCGUAUUUUCAAGCCCAGCGUGAAAAUACCGCGCGCAGUAAUGAAAUGAAAUGUAAUUU ...........((((((((..((.((......)).))....--.......(((((((....(.(((((((((.((...)).))))))))).).)))))))..)))))))).......... ( -27.40) >DroEre_CAF1 79402 115 - 1 CUAUUUUUAACUCGUUUCGCCUGCAGG--UCUCUUUAUUUU---UUUCCCGCUGCGCAUUCUUGUAUUUCCAAGCCCAGCGUGAAAAUACCGCGCACAGUAAUGAAAUGAAAUGUAAUUU ...........(((((((((.(((.((--(..........(---((((.(((((.((....(((......))))).))))).))))).)))..)))..))...))))))).......... ( -21.70) >DroYak_CAF1 77202 115 - 1 CUAUUUUUAACUCGUUUCGCCUGCAGU--GCUCUUUAUUUU---UUUCCCGCUGCGCAUUCUUGUAUUUCCAAGCCCAGCGUGAAAAUACCGCGCACAGUAAUGAAAUGAAAUGUAAUUU ...........((((((((..(((.((--((.(..((((((---(....(((((.((....(((......))))).))))).)))))))..).)))).))).)))))))).......... ( -26.80) >consensus CUAUUUUUAACUCGUUUCGCCUGCAGUCUCCUCUUUAUUUU___UUUCCCGCUGCGCAUUCCCGUAUUUUCAAGCCCAGCGUGAAAAUACCGCGCGCAGUAAUGAAAUGAAAUGUAAUUU ...........((((((((....)..........................(((((((......(((((((((.((...)).)))))))))...)))))))...))))))).......... (-22.16 = -22.96 + 0.80)

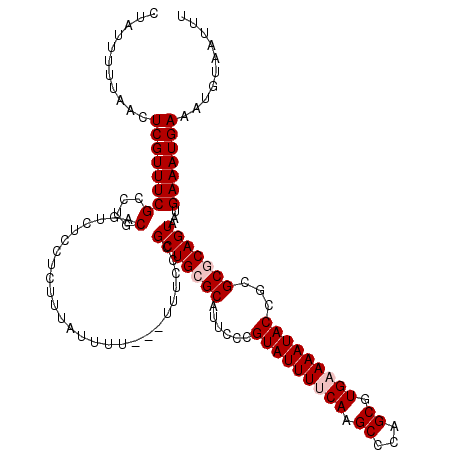
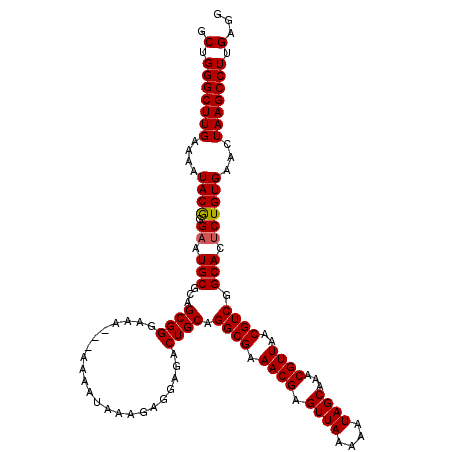
| Location | 12,956,040 – 12,956,160 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.67 |
| Mean single sequence MFE | -29.56 |
| Consensus MFE | -25.89 |
| Energy contribution | -25.85 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.947588 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12956040 120 + 23771897 GCUGGGCUUGAAAAUACGGGAAUGCGCAGCGGGAAAAUAAAAAUAAAGAGGAGACUGCAGGCGAAACGAGUUAAAAAUAGCAAACGUUAACGUCGGCACUCUGUGAACUAAGCCUUGAGG .(.(((((((....((((((..((((((((......................).)))).((((.((((.((((....))))...))))..)))).))).))))))...))))))).)... ( -26.95) >DroSec_CAF1 73242 117 + 1 GCUGGGCUUGAAAAUACGGGAAUGCGCAGCGGGAAA---AAAAUAAAGAGGAGACUGCAGGCGAAACGAGUUAAAAAUAGCAAACGUUAACGUCGGCACUCUGUGAACUAAGCCUUGAGG .(.(((((((....((((((..((((((((......---.............).)))).((((.((((.((((....))))...))))..)))).))).))))))...))))))).)... ( -27.11) >DroSim_CAF1 78064 118 + 1 GCUGGGCUUGAAAAUACGGGAAUGCGCAGCGGGAAAA--AAAAUGAAGAGGAGACUGCAGGCGAAACGAGUUAAAAAUAGCAAACGUUAACGUCGGCACGCUGUGAACUAAGCCUUGAGG .(.(((((((......((....))(((((((......--.........((....))((.((((.((((.((((....))))...))))..)))).)).)))))))...))))))).)... ( -31.10) >DroEre_CAF1 79442 115 + 1 GCUGGGCUUGGAAAUACAAGAAUGCGCAGCGGGAAA---AAAAUAAAGAGA--CCUGCAGGCGAAACGAGUUAAAAAUAGCAAACGUUAACGUCGGCACUCUGUGAACUAAGCCUUGAGG .(.((((((((...((((.((.(((...(((((...---............--))))).((((.((((.((((....))))...))))..)))).))).))))))..)))))))).)... ( -32.26) >DroYak_CAF1 77242 115 + 1 GCUGGGCUUGGAAAUACAAGAAUGCGCAGCGGGAAA---AAAAUAAAGAGC--ACUGCAGGCGAAACGAGUUAAAAAUAGCAAACGUUAACGUCGGCACUCUGUGAACUAAGCCUUGAGG .(.((((((((...((((.((.(((((((.(.....---...........)--.)))).((((.((((.((((....))))...))))..)))).))).))))))..)))))))).)... ( -30.39) >consensus GCUGGGCUUGAAAAUACGGGAAUGCGCAGCGGGAAA___AAAAUAAAGAGGAGACUGCAGGCGAAACGAGUUAAAAAUAGCAAACGUUAACGUCGGCACUCUGUGAACUAAGCCUUGAGG .(.(((((((....((((.((.(((...((((......................)))).((((.((((.((((....))))...))))..)))).))).))))))...))))))).)... (-25.89 = -25.85 + -0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:23:46 2006