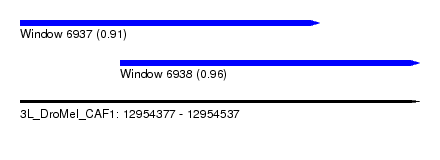
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,954,377 – 12,954,537 |
| Length | 160 |
| Max. P | 0.961804 |

| Location | 12,954,377 – 12,954,497 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.31 |
| Mean single sequence MFE | -38.86 |
| Consensus MFE | -33.58 |
| Energy contribution | -34.06 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.909474 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12954377 120 + 23771897 GGGCGUGGCAGGGCGUGUCUUGACCGGGCUAGUAAACUAUGCAAACAAAUAGCCAACUUAUUUUGGGGGGCCCCGAAGUAAAUUGUACAGAGCAGCGCAAAAUUCGUGCUGCAAAGCAAC .(((.((((((((....))))).))).))).........(((...........(((.((((((((((....)))))))))).)))......(((((((.......)))))))...))).. ( -43.30) >DroSec_CAF1 71589 119 + 1 GGGCGUGGCAGGGCGUGUCUUGACCGGGCUAGUAAACUAUGCAAACAAAUAGCCAACUUAUUUUGG-GGGCCCCGAAGUAAAUUGUACAGAGCAGCGCAGAAUUCGUGCUGCAAAGCAAC .(((.((((((((....))))).))).))).........(((...........(((.(((((((((-(...)))))))))).)))......(((((((.......)))))))...))).. ( -40.30) >DroSim_CAF1 76421 119 + 1 GGGAGUGGCAGGGCGUGUCUUGACCGGGCUAGUAAACUAUGCAAACAAAUAGCCAACUUAUUUCGG-GGGCCCCGAAGUAAAUUGUACAGAGCAGCGCAGAAUUCGUGCUGCAAAGCAAC .....((((((((....))))).))).(((.(((..((((........)))).(((.(((((((((-(...)))))))))).))))))...(((((((.......)))))))..)))... ( -38.00) >DroEre_CAF1 77665 119 + 1 GGGCGUGGCAGGGCGUGUCUUGACCGGGCUAGUAAACUAUGCAAACAAAUAGCCAACUUAUUUUGG-GCGCACCGAAGUAAAUUGUACAGACUAGCGCAGAAUUCGUGCUGCAAAGCAAC ..((...((((.(((..(((.....(.((((((...((((........)))).(((.(((((((((-.....))))))))).))).....)))))).))))...))).))))...))... ( -37.70) >DroYak_CAF1 75470 119 + 1 GGGCGGGGCAGGGCGUGUCUUGACCGGGCUAGUAAACUAUGCAAACAAAUAGCCAACUUAUUUUGG-GGGUCCGGAAGUAAAUUGUACAGAGCAGCGCAGAAUUCGUGCUGCAAAGCAAC ...(((((((.....))))))).(((((((......((((........))))((((......))))-.)))))))..(((.....)))...(((((((.......)))))))........ ( -35.00) >consensus GGGCGUGGCAGGGCGUGUCUUGACCGGGCUAGUAAACUAUGCAAACAAAUAGCCAACUUAUUUUGG_GGGCCCCGAAGUAAAUUGUACAGAGCAGCGCAGAAUUCGUGCUGCAAAGCAAC .(((.((((((((....))))).))).))).........(((...........(((.(((((((((......))))))))).)))......(((((((.......)))))))...))).. (-33.58 = -34.06 + 0.48)


| Location | 12,954,417 – 12,954,537 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.30 |
| Mean single sequence MFE | -33.58 |
| Consensus MFE | -27.24 |
| Energy contribution | -27.32 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.961804 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12954417 120 + 23771897 GCAAACAAAUAGCCAACUUAUUUUGGGGGGCCCCGAAGUAAAUUGUACAGAGCAGCGCAAAAUUCGUGCUGCAAAGCAACAGCUGUCGGAAAACUCAAAGUCUGGUCUGUAAAAUAUCAG .............(((.((((((((((....)))))))))).)))(((((((((((((.......))))))).......(((((....((....))..)).))).))))))......... ( -36.30) >DroSec_CAF1 71629 119 + 1 GCAAACAAAUAGCCAACUUAUUUUGG-GGGCCCCGAAGUAAAUUGUACAGAGCAGCGCAGAAUUCGUGCUGCAAAGCAACAGCUGUCGGAAAACUCAAAGUCUGGUCUGUAAAAUAUCAG .............(((.(((((((((-(...)))))))))).)))(((((((((((((.......))))))).......(((((....((....))..)).))).))))))......... ( -33.30) >DroSim_CAF1 76461 119 + 1 GCAAACAAAUAGCCAACUUAUUUCGG-GGGCCCCGAAGUAAAUUGUACAGAGCAGCGCAGAAUUCGUGCUGCAAAGCAACAGCUGUCGGAAAACUCAAAGUCUGGUCUGUAAAAUAUCAG .............(((.(((((((((-(...)))))))))).)))(((((((((((((.......))))))).......(((((....((....))..)).))).))))))......... ( -35.40) >DroEre_CAF1 77705 117 + 1 GCAAACAAAUAGCCAACUUAUUUUGG-GCGCACCGAAGUAAAUUGUACAGACUAGCGCAGAAUUCGUGCUGCAAAGCAACAGCUGUCGGACAACUCAAAG--UGGUCUGUAAAAUAUCGG .............(((.(((((((((-.....))))))))).)))(((((((((.(((((......((((....))))....))).))((....))....--)))))))))......... ( -29.70) >DroYak_CAF1 75510 117 + 1 GCAAACAAAUAGCCAACUUAUUUUGG-GGGUCCGGAAGUAAAUUGUACAGAGCAGCGCAGAAUUCGUGCUGCAAAGCAACAGCUGUCGGAAAACUCAAAG--UGGUCUGUAAAAUAUCAG ........(((((((.....((((((-(..((((..(((...((((.....(((((((.......)))))))...))))..)))..))))...)))))))--))).)))).......... ( -33.20) >consensus GCAAACAAAUAGCCAACUUAUUUUGG_GGGCCCCGAAGUAAAUUGUACAGAGCAGCGCAGAAUUCGUGCUGCAAAGCAACAGCUGUCGGAAAACUCAAAGUCUGGUCUGUAAAAUAUCAG .............(((.(((((((((......))))))))).)))(((((((((((((.......)))))))..(((....))).....................))))))......... (-27.24 = -27.32 + 0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:23:34 2006