
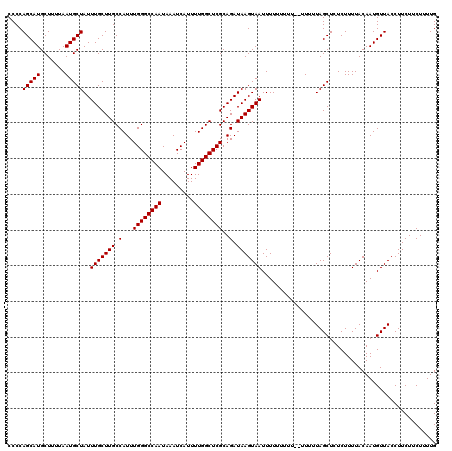
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,954,149 – 12,954,308 |
| Length | 159 |
| Max. P | 0.999700 |

| Location | 12,954,149 – 12,954,268 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.12 |
| Mean single sequence MFE | -18.76 |
| Consensus MFE | -18.70 |
| Energy contribution | -18.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.57 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.22 |
| SVM RNA-class probability | 0.990640 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12954149 119 - 23771897 CCCCAGCAUGCUUUUAAUGCUAUUUGCUUGCCAUUUGGGCCAAUAAAUCAUUUUGGCUCGCAGAUAAGUAAUUUUUUUUUU-UUUUUAGCUCUCUUUUACAAUGUUACCUUCUUCUUUUG ....(((((.......)))))..(((((((.(...((((((((.........))))))))..).)))))))..........-...................................... ( -18.70) >DroSec_CAF1 71362 118 - 1 CCCCAGCAUGCUUUUAAUGCUAUUUGCUUGCCAUUUGGGCCAAUAAAUCAUUUUGGCUCGCAGAUAAGUAAUUUUUUUUU--UUUUUAGCUCUCUUUUACAAUGUUACCUUCUUCUUUUG ....(((((.......)))))..(((((((.(...((((((((.........))))))))..).))))))).........--...................................... ( -18.70) >DroSim_CAF1 76192 120 - 1 CCCCAGCAUGCUUUUAAUGCUAUUUGCUUGCCAUUUGGGCCAAUAAAUCAUUUUGGCUCGCAGAUAAGUAAUUUUUUUUUUCUUUUUAGCUCUCUUUUACAAUGUUACCUUCUUCUUUUG ....(((((.......)))))..(((((((.(...((((((((.........))))))))..).)))))))................................................. ( -18.70) >DroEre_CAF1 77436 112 - 1 CCCCAGCAUGCUUUUAAUGCUAUUUGCUUGCCAUUUGGGCCAAUAAAUCAUUUUGGCUCGCAGAUAAGUAAUUUU--------UUUUAGCUCUCUUUUACAAUGUUACCUUCUUCUUUUG ....(((((.........((((.(((((((.(...((((((((.........))))))))..).)))))))....--------...))))...........))))).............. ( -18.85) >DroYak_CAF1 75250 112 - 1 CCCCAGCAUGCUUUUAAUGCUAUUUGCUUGCCAUUUGGGCCAAUAAAUCAUUUUGGCUCGCAGAUAAGUAAUUUU--------UUUUAGCUCUCUUUUACAAUGUUACCUUCUUCUUUUG ....(((((.........((((.(((((((.(...((((((((.........))))))))..).)))))))....--------...))))...........))))).............. ( -18.85) >consensus CCCCAGCAUGCUUUUAAUGCUAUUUGCUUGCCAUUUGGGCCAAUAAAUCAUUUUGGCUCGCAGAUAAGUAAUUUUUUUUU__UUUUUAGCUCUCUUUUACAAUGUUACCUUCUUCUUUUG ....(((((.......)))))..(((((((.(...((((((((.........))))))))..).)))))))................................................. (-18.70 = -18.70 + -0.00)


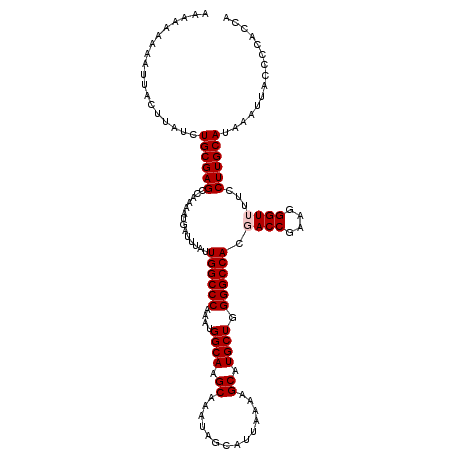
| Location | 12,954,188 – 12,954,308 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.31 |
| Mean single sequence MFE | -28.62 |
| Consensus MFE | -27.36 |
| Energy contribution | -27.76 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.96 |
| SVM decision value | 3.91 |
| SVM RNA-class probability | 0.999700 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12954188 120 + 23771897 AAAAAAAAAUUACUUAUCUGCGAGCCAAAAUGAUUUAUUGGCCCAAAUGGCAAGCAAAUAGCAUUAAAAGCAUGCUGGGGCCACGACCGAAGGGUUUUCCUUGCAUAAAUUACCCCACCA ..................((((((..............((((((....((((.((..............)).)))).)))))).((((....))))...))))))............... ( -29.04) >DroSec_CAF1 71400 120 + 1 AAAAAAAAAUUACUUAUCUGCGAGCCAAAAUGAUUUAUUGGCCCAAAUGGCAAGCAAAUAGCAUUAAAAGCAUGCUGGGGCCACGACCGAAUGGUUUUCCUUGCAUAAAUUACCCCACCA ..................((((((..(((((.((((..((((((....((((.((..............)).)))).)))))).....)))).))))).))))))............... ( -26.94) >DroSim_CAF1 76232 120 + 1 AAAAAAAAAUUACUUAUCUGCGAGCCAAAAUGAUUUAUUGGCCCAAAUGGCAAGCAAAUAGCAUUAAAAGCAUGCUGGGGCCACGACCGAAGGGUUUUCCUUGCAUAAAUUACCCCACCA ..................((((((..............((((((....((((.((..............)).)))).)))))).((((....))))...))))))............... ( -29.04) >DroEre_CAF1 77473 114 + 1 -----AAAAUUACUUAUCUGCGAGCCAAAAUGAUUUAUUGGCCCAAAUGGCAAGCAAAUAGCAUUAAAAGCAUGCUGGGGCCACGACCGAAGGGU-UUCCUUGCAUAAAUUACCCCACCA -----.............((((((..............((((((....((((.((..............)).)))).)))))).((((....)))-)..))))))............... ( -29.04) >DroYak_CAF1 75287 114 + 1 -----AAAAUUACUUAUCUGCGAGCCAAAAUGAUUUAUUGGCCCAAAUGGCAAGCAAAUAGCAUUAAAAGCAUGCUGGGGCCACGACCGAAGGGU-UUCCUUGCAUAAAUUACCCCACAA -----.............((((((..............((((((....((((.((..............)).)))).)))))).((((....)))-)..))))))............... ( -29.04) >consensus AAAAAAAAAUUACUUAUCUGCGAGCCAAAAUGAUUUAUUGGCCCAAAUGGCAAGCAAAUAGCAUUAAAAGCAUGCUGGGGCCACGACCGAAGGGUUUUCCUUGCAUAAAUUACCCCACCA ..................((((((..............((((((....((((.((..............)).)))).)))))).((((....))))...))))))............... (-27.36 = -27.76 + 0.40)


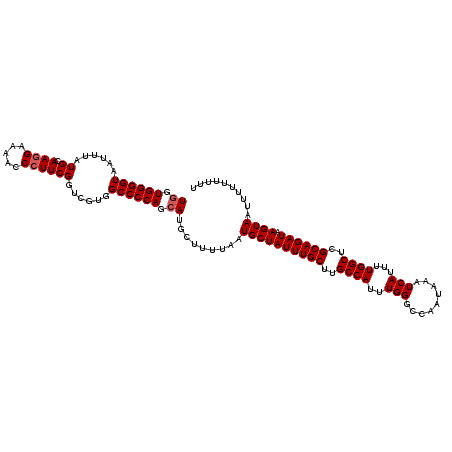
| Location | 12,954,188 – 12,954,308 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.31 |
| Mean single sequence MFE | -33.26 |
| Consensus MFE | -31.86 |
| Energy contribution | -32.26 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.76 |
| SVM RNA-class probability | 0.996845 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12954188 120 - 23771897 UGGUGGGGUAAUUUAUGCAAGGAAAACCCUUCGGUCGUGGCCCCAGCAUGCUUUUAAUGCUAUUUGCUUGCCAUUUGGGCCAAUAAAUCAUUUUGGCUCGCAGAUAAGUAAUUUUUUUUU ((.((((((......((.((((.....))))))......)))))).)).........((((((((((..((((..(((.........)))...))))..)))))).)))).......... ( -34.30) >DroSec_CAF1 71400 120 - 1 UGGUGGGGUAAUUUAUGCAAGGAAAACCAUUCGGUCGUGGCCCCAGCAUGCUUUUAAUGCUAUUUGCUUGCCAUUUGGGCCAAUAAAUCAUUUUGGCUCGCAGAUAAGUAAUUUUUUUUU ((.((((((......((...((....))...))......)))))).)).........((((((((((..((((..(((.........)))...))))..)))))).)))).......... ( -30.30) >DroSim_CAF1 76232 120 - 1 UGGUGGGGUAAUUUAUGCAAGGAAAACCCUUCGGUCGUGGCCCCAGCAUGCUUUUAAUGCUAUUUGCUUGCCAUUUGGGCCAAUAAAUCAUUUUGGCUCGCAGAUAAGUAAUUUUUUUUU ((.((((((......((.((((.....))))))......)))))).)).........((((((((((..((((..(((.........)))...))))..)))))).)))).......... ( -34.30) >DroEre_CAF1 77473 114 - 1 UGGUGGGGUAAUUUAUGCAAGGAA-ACCCUUCGGUCGUGGCCCCAGCAUGCUUUUAAUGCUAUUUGCUUGCCAUUUGGGCCAAUAAAUCAUUUUGGCUCGCAGAUAAGUAAUUUU----- ((.((((((......((.((((..-..))))))......)))))).)).........((((((((((..((((..(((.........)))...))))..)))))).)))).....----- ( -34.30) >DroYak_CAF1 75287 114 - 1 UUGUGGGGUAAUUUAUGCAAGGAA-ACCCUUCGGUCGUGGCCCCAGCAUGCUUUUAAUGCUAUUUGCUUGCCAUUUGGGCCAAUAAAUCAUUUUGGCUCGCAGAUAAGUAAUUUU----- ...((((((......((.((((..-..))))))......))))))((((.......))))(((((((..((((..(((.........)))...))))..))))))).........----- ( -33.10) >consensus UGGUGGGGUAAUUUAUGCAAGGAAAACCCUUCGGUCGUGGCCCCAGCAUGCUUUUAAUGCUAUUUGCUUGCCAUUUGGGCCAAUAAAUCAUUUUGGCUCGCAGAUAAGUAAUUUUUUUUU ((.((((((......((.((((.....))))))......)))))).)).........((((((((((..((((..(((.........)))...))))..)))))).)))).......... (-31.86 = -32.26 + 0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:23:32 2006