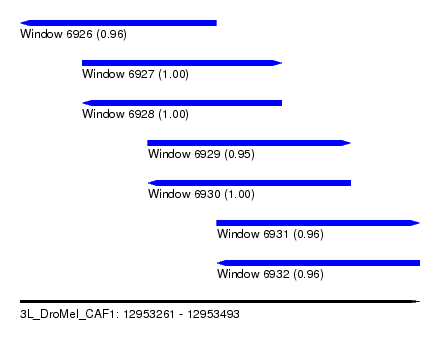
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,953,261 – 12,953,493 |
| Length | 232 |
| Max. P | 0.999992 |

| Location | 12,953,261 – 12,953,375 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.67 |
| Mean single sequence MFE | -37.38 |
| Consensus MFE | -33.70 |
| Energy contribution | -33.74 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.958968 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12953261 114 - 23771897 GUUGGUUUAAUGAUGUCACUUGGUGGUUGGCUGCUCAAUUCUGCCCGCCGGCAAAGGAGCAUUAUCAUUAAUGUAUUC------GUCUGCUCACAGUAGGCAAUCAGGCGUCGUGUAGUU .........((((((((((((((((((....(((((.....((((....))))...)))))..)))))))).))....------((((((.....)))))).....))))))))...... ( -34.80) >DroSec_CAF1 70468 114 - 1 GUUGGUUUAAUGAUGUCACUUGGUGGUUGGCUGCUCCAUUCUGCCCGCCGGCAAAGGAGCAUUAUCAUUAAUGUAUUC------GUCUGCUCACAGUAGGCAAUCAGGCGUCGUGUAGUU .........((((((((((((((((((....((((((.((.((((....))))))))))))..)))))))).))....------((((((.....)))))).....))))))))...... ( -37.30) >DroSim_CAF1 75319 114 - 1 GUUGGUUUAAUGAUGUCACUUGGUGGUUGGCUGCUCAAUUCUGCCCGCCGGCAAAGGAGCAUUAUCAUUAAUGUAUUC------GUCUGCUCACAGUAGGCAAUCAGGCGUCGUGUAGUU .........((((((((((((((((((....(((((.....((((....))))...)))))..)))))))).))....------((((((.....)))))).....))))))))...... ( -34.80) >DroEre_CAF1 76549 120 - 1 GUUAGUUUAAUGAUGUCACUUGGUGGUUGGCUGCUCAAUUCUGCCCGCCGGCAAAGGAGCAUUAUCAUUAAUGUAUUCCUGCCUGUCUGCUCACAGUAGGCAAUCAGGCGUCGUGUAGUU .........((((((((..(((.((((.(((...........))).)))).)))((((((((((....))))))..))))(((((.(((....)))))))).....))))))))...... ( -39.80) >DroYak_CAF1 74313 120 - 1 GUUAGUUUAAUGAUGUCACUUGGUGGUUGGCUGCUCAAUUUUGCCCGCCGGCAAAGGAGCAUUAUCAUUAAUGUAUUCCUGGCUGUCUGCUCGCAGUUGGCAAUCAGGCAUCGUGUAGUU .........((((((((...((((.((..(((((......(((((....)))))((((((((((....))))))..))))(((.....))).)))))..)).))))))))))))...... ( -40.20) >consensus GUUGGUUUAAUGAUGUCACUUGGUGGUUGGCUGCUCAAUUCUGCCCGCCGGCAAAGGAGCAUUAUCAUUAAUGUAUUC______GUCUGCUCACAGUAGGCAAUCAGGCGUCGUGUAGUU .........((((((((((((((((((....(((((.....((((....))))...)))))..)))))))).))..........((((((.....)))))).....))))))))...... (-33.70 = -33.74 + 0.04)

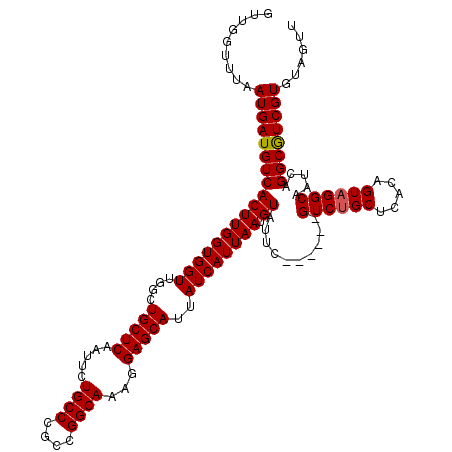

| Location | 12,953,297 – 12,953,413 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.77 |
| Mean single sequence MFE | -32.90 |
| Consensus MFE | -31.32 |
| Energy contribution | -31.36 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.51 |
| Structure conservation index | 0.95 |
| SVM decision value | 5.26 |
| SVM RNA-class probability | 0.999981 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12953297 116 + 23771897 --GAAUACAUUAAUGAUAAUGCUCCUUUGCCGGCGGGCAGAAUUGAGCAGCCAACCACCAAGUGACAUCAUUAAACCAACUGCCAAAGUAAAUCAUAUAU--GUAUGAAACUUUGGGCAA --.......((((((((..(((((.((((((....))))))...)))))......(((...)))..))))))))......(((((((((...(((((...--.))))).))))).)))). ( -31.60) >DroSec_CAF1 70504 116 + 1 --GAAUACAUUAAUGAUAAUGCUCCUUUGCCGGCGGGCAGAAUGGAGCAGCCAACCACCAAGUGACAUCAUUAAACCAACUGCCAAAGUAAAUCAUAUGU--GUAUGAAACUUUAGGCAA --.......((((((((..((((((((((((....))))))..))))))......(((...)))..))))))))......(((((((((...(((((...--.))))).))))).)))). ( -35.00) >DroSim_CAF1 75355 116 + 1 --GAAUACAUUAAUGAUAAUGCUCCUUUGCCGGCGGGCAGAAUUGAGCAGCCAACCACCAAGUGACAUCAUUAAACCAACUGCCAAAGUAAAUCAUAUAU--GUAUGAAACUUUGGGCAA --.......((((((((..(((((.((((((....))))))...)))))......(((...)))..))))))))......(((((((((...(((((...--.))))).))))).)))). ( -31.60) >DroEre_CAF1 76589 118 + 1 AGGAAUACAUUAAUGAUAAUGCUCCUUUGCCGGCGGGCAGAAUUGAGCAGCCAACCACCAAGUGACAUCAUUAAACUAACUGCCAAAGUAAAUCAUAUAU--GUAUGAAACUUUGGCCAA .........((((((((..(((((.((((((....))))))...)))))......(((...)))..)))))))).......((((((((...(((((...--.))))).))))))))... ( -33.70) >DroYak_CAF1 74353 120 + 1 AGGAAUACAUUAAUGAUAAUGCUCCUUUGCCGGCGGGCAAAAUUGAGCAGCCAACCACCAAGUGACAUCAUUAAACUAACUGCCAAAGUAAAUCAUAUAUGUGUAUGAAACUUUGGGCAU .........((((((((..(((((.((((((....))))))...)))))......(((...)))..))))))))......(((((((((...((((((....)))))).))))).)))). ( -32.60) >consensus __GAAUACAUUAAUGAUAAUGCUCCUUUGCCGGCGGGCAGAAUUGAGCAGCCAACCACCAAGUGACAUCAUUAAACCAACUGCCAAAGUAAAUCAUAUAU__GUAUGAAACUUUGGGCAA .........((((((((..(((((.((((((....))))))...)))))......(((...)))..))))))))......(((((((((...((((((....)))))).))))))).)). (-31.32 = -31.36 + 0.04)


| Location | 12,953,297 – 12,953,413 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.77 |
| Mean single sequence MFE | -35.16 |
| Consensus MFE | -34.22 |
| Energy contribution | -33.82 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.97 |
| SVM decision value | 5.25 |
| SVM RNA-class probability | 0.999980 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12953297 116 - 23771897 UUGCCCAAAGUUUCAUAC--AUAUAUGAUUUACUUUGGCAGUUGGUUUAAUGAUGUCACUUGGUGGUUGGCUGCUCAAUUCUGCCCGCCGGCAAAGGAGCAUUAUCAUUAAUGUAUUC-- .((((.(((((.(((((.--...)))))...)))))))))((((((...(((((((..((((.((((.(((...........))).)))).).)))..))))))).))))))......-- ( -33.90) >DroSec_CAF1 70504 116 - 1 UUGCCUAAAGUUUCAUAC--ACAUAUGAUUUACUUUGGCAGUUGGUUUAAUGAUGUCACUUGGUGGUUGGCUGCUCCAUUCUGCCCGCCGGCAAAGGAGCAUUAUCAUUAAUGUAUUC-- (((((.(((((.(((((.--...)))))...)))))))))).....((((((((((((((....)).))))((((((.((.((((....))))))))))))..)))))))).......-- ( -35.40) >DroSim_CAF1 75355 116 - 1 UUGCCCAAAGUUUCAUAC--AUAUAUGAUUUACUUUGGCAGUUGGUUUAAUGAUGUCACUUGGUGGUUGGCUGCUCAAUUCUGCCCGCCGGCAAAGGAGCAUUAUCAUUAAUGUAUUC-- .((((.(((((.(((((.--...)))))...)))))))))((((((...(((((((..((((.((((.(((...........))).)))).).)))..))))))).))))))......-- ( -33.90) >DroEre_CAF1 76589 118 - 1 UUGGCCAAAGUUUCAUAC--AUAUAUGAUUUACUUUGGCAGUUAGUUUAAUGAUGUCACUUGGUGGUUGGCUGCUCAAUUCUGCCCGCCGGCAAAGGAGCAUUAUCAUUAAUGUAUUCCU ...((((((((.(((((.--...)))))...)))))))).((((((...(((((((..((((.((((.(((...........))).)))).).)))..))))))).))))))........ ( -37.20) >DroYak_CAF1 74353 120 - 1 AUGCCCAAAGUUUCAUACACAUAUAUGAUUUACUUUGGCAGUUAGUUUAAUGAUGUCACUUGGUGGUUGGCUGCUCAAUUUUGCCCGCCGGCAAAGGAGCAUUAUCAUUAAUGUAUUCCU .((((.(((((.(((((......)))))...)))))))))......((((((((((((((....)).))))(((((...((((((....)))))).)))))..))))))))......... ( -35.40) >consensus UUGCCCAAAGUUUCAUAC__AUAUAUGAUUUACUUUGGCAGUUGGUUUAAUGAUGUCACUUGGUGGUUGGCUGCUCAAUUCUGCCCGCCGGCAAAGGAGCAUUAUCAUUAAUGUAUUC__ .((.(((((((.(((((......)))))...)))))))))((((((...(((((((..((((.((((.(((...........))).)))).).)))..))))))).))))))........ (-34.22 = -33.82 + -0.40)



| Location | 12,953,335 – 12,953,453 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.95 |
| Mean single sequence MFE | -27.46 |
| Consensus MFE | -23.60 |
| Energy contribution | -23.76 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.946730 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12953335 118 + 23771897 AAUUGAGCAGCCAACCACCAAGUGACAUCAUUAAACCAACUGCCAAAGUAAAUCAUAUAU--GUAUGAAACUUUGGGCAACAGUUCUGGGUAUAACAAUAAUCUUUUGGUAGGUGGGAGA ..........((.(((((((((.((.....(((.(((((((((((((((...(((((...--.))))).)))))))....)))))...))).)))......)).)))))).))).))... ( -28.50) >DroSec_CAF1 70542 118 + 1 AAUGGAGCAGCCAACCACCAAGUGACAUCAUUAAACCAACUGCCAAAGUAAAUCAUAUGU--GUAUGAAACUUUAGGCAACAAUUCUGGGUAUAACAAUAAUCUUUUGGUAGGUGGGAGA ..(((.....))).(((((.((((....))))..(((((.(((((((((...(((((...--.))))).))))).)))).......((.......))........))))).))))).... ( -27.80) >DroSim_CAF1 75393 118 + 1 AAUUGAGCAGCCAACCACCAAGUGACAUCAUUAAACCAACUGCCAAAGUAAAUCAUAUAU--GUAUGAAACUUUGGGCAACAAUUCGGGGUAUAACAAUAAUCUUUUGGUAGGUGGGAGA ..............(((((.((((....))))..(((((...(((((((...(((((...--.))))).)))))))((..(.....)..))..............))))).))))).... ( -27.10) >DroEre_CAF1 76629 118 + 1 AAUUGAGCAGCCAACCACCAAGUGACAUCAUUAAACUAACUGCCAAAGUAAAUCAUAUAU--GUAUGAAACUUUGGCCAACAGUUUUGGGUACAACAAUAAUCUUUUGGUAGGUGGGAGA ..............(((((..((...(((...(((((....((((((((...(((((...--.))))).))))))))....)))))..)))...))....(((....))).))))).... ( -29.10) >DroYak_CAF1 74393 120 + 1 AAUUGAGCAGCCAACCACCAAGUGACAUCAUUAAACUAACUGCCAAAGUAAAUCAUAUAUGUGUAUGAAACUUUGGGCAUCAGUUUUGGGUACAACAAUAAUCUUUUGGUAUGUGGGAGA ..........(((...((((((.((............((((((((((((...((((((....)))))).)))))))....)))))(((....)))......)).))))))...))).... ( -24.80) >consensus AAUUGAGCAGCCAACCACCAAGUGACAUCAUUAAACCAACUGCCAAAGUAAAUCAUAUAU__GUAUGAAACUUUGGGCAACAGUUCUGGGUAUAACAAUAAUCUUUUGGUAGGUGGGAGA ..............(((((.((((....))))..(((((.(((((((((...((((((....)))))).))))))).))........((((.........)))).))))).))))).... (-23.60 = -23.76 + 0.16)


| Location | 12,953,335 – 12,953,453 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.95 |
| Mean single sequence MFE | -33.24 |
| Consensus MFE | -32.30 |
| Energy contribution | -32.26 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.52 |
| Structure conservation index | 0.97 |
| SVM decision value | 5.70 |
| SVM RNA-class probability | 0.999992 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12953335 118 - 23771897 UCUCCCACCUACCAAAAGAUUAUUGUUAUACCCAGAACUGUUGCCCAAAGUUUCAUAC--AUAUAUGAUUUACUUUGGCAGUUGGUUUAAUGAUGUCACUUGGUGGUUGGCUGCUCAAUU ....(((.(((((((..(((.((..(((.(((...(((((....(((((((.(((((.--...)))))...))))))))))))))).)))..)))))..))))))).))).......... ( -35.70) >DroSec_CAF1 70542 118 - 1 UCUCCCACCUACCAAAAGAUUAUUGUUAUACCCAGAAUUGUUGCCUAAAGUUUCAUAC--ACAUAUGAUUUACUUUGGCAGUUGGUUUAAUGAUGUCACUUGGUGGUUGGCUGCUCCAUU ....(((.(((((((..(((.((..(((...((((.....(((((.(((((.(((((.--...)))))...))))))))))))))..)))..)))))..))))))).))).......... ( -32.70) >DroSim_CAF1 75393 118 - 1 UCUCCCACCUACCAAAAGAUUAUUGUUAUACCCCGAAUUGUUGCCCAAAGUUUCAUAC--AUAUAUGAUUUACUUUGGCAGUUGGUUUAAUGAUGUCACUUGGUGGUUGGCUGCUCAAUU ....(((.(((((((..(((.((..(((.(((...(((((....(((((((.(((((.--...)))))...))))))))))))))).)))..)))))..))))))).))).......... ( -33.40) >DroEre_CAF1 76629 118 - 1 UCUCCCACCUACCAAAAGAUUAUUGUUGUACCCAAAACUGUUGGCCAAAGUUUCAUAC--AUAUAUGAUUUACUUUGGCAGUUAGUUUAAUGAUGUCACUUGGUGGUUGGCUGCUCAAUU ....(((.(((((((..(((.((..(((.......(((((...((((((((.(((((.--...)))))...))))))))...))))))))..)))))..))))))).))).......... ( -34.51) >DroYak_CAF1 74393 120 - 1 UCUCCCACAUACCAAAAGAUUAUUGUUGUACCCAAAACUGAUGCCCAAAGUUUCAUACACAUAUAUGAUUUACUUUGGCAGUUAGUUUAAUGAUGUCACUUGGUGGUUGGCUGCUCAAUU ....(((..((((((..(((.((..(((.......(((((((..(((((((.(((((......)))))...)))))))..))))))))))..)))))..))))))..))).......... ( -29.91) >consensus UCUCCCACCUACCAAAAGAUUAUUGUUAUACCCAGAACUGUUGCCCAAAGUUUCAUAC__AUAUAUGAUUUACUUUGGCAGUUGGUUUAAUGAUGUCACUUGGUGGUUGGCUGCUCAAUU ....(((.(((((((..(((.((..(((.(((...(((((....(((((((.(((((......)))))...))))))))))))))).)))..)))))..))))))).))).......... (-32.30 = -32.26 + -0.04)



| Location | 12,953,375 – 12,953,493 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.10 |
| Mean single sequence MFE | -30.44 |
| Consensus MFE | -23.92 |
| Energy contribution | -24.52 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958512 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12953375 118 + 23771897 UGCCAAAGUAAAUCAUAUAU--GUAUGAAACUUUGGGCAACAGUUCUGGGUAUAACAAUAAUCUUUUGGUAGGUGGGAGAAACCAGGCAAAUAAAUUUAUGGCAGUUGAACUCUUGGCAA (((((((((...(((((...--.))))).))))).))))...(((..((((.((((.((((...((((.(.(((.......))).).)))).....))))....)))).))))..))).. ( -25.40) >DroSec_CAF1 70582 118 + 1 UGCCAAAGUAAAUCAUAUGU--GUAUGAAACUUUAGGCAACAAUUCUGGGUAUAACAAUAAUCUUUUGGUAGGUGGGAGAACCCAGGCAAAUAAAUUUAUGCCAGUUGAACUCUUGGCAA (((((((((...(((..((.--(((((((......(....)...(((((((.(..((...(((....)))...))..)..)))))))........)))))))))..)))))).)))))). ( -30.40) >DroSim_CAF1 75433 118 + 1 UGCCAAAGUAAAUCAUAUAU--GUAUGAAACUUUGGGCAACAAUUCGGGGUAUAACAAUAAUCUUUUGGUAGGUGGGAGAACCCAGGCAAAUAAAUUUAUGCCAGUUGAACUCUUGGCAA (((((((((...(((((...--.))))).)))))(..((((.....(((...(..((...(((....)))...))..)...))).((((.(......).)))).))))..)....)))). ( -29.00) >DroEre_CAF1 76669 118 + 1 UGCCAAAGUAAAUCAUAUAU--GUAUGAAACUUUGGCCAACAGUUUUGGGUACAACAAUAAUCUUUUGGUAGGUGGGAGAACCCAUCCAAAUAAAUUUAUGGCAGUUGAACUCUCGACAA .((((((((...(((((...--.))))).)))))))).....(((..((((.((((.((((...(((((...(((((....)))))))))).....))))....)))).))))..))).. ( -33.80) >DroYak_CAF1 74433 120 + 1 UGCCAAAGUAAAUCAUAUAUGUGUAUGAAACUUUGGGCAUCAGUUUUGGGUACAACAAUAAUCUUUUGGUAUGUGGGAGAACCCAUCCAAAUAAAUUUAUGGCAGUUGAACUCUUGACAA (((((((((...((((((....)))))).))))).))))...(((..((((.((((.((((...(((((...(((((....)))))))))).....))))....)))).))))..))).. ( -33.60) >consensus UGCCAAAGUAAAUCAUAUAU__GUAUGAAACUUUGGGCAACAGUUCUGGGUAUAACAAUAAUCUUUUGGUAGGUGGGAGAACCCAGGCAAAUAAAUUUAUGGCAGUUGAACUCUUGGCAA (((((((((...((((((....)))))).))))))).))...(((..((((.((((.................((((....))))((((.(......).)))).)))).))))..))).. (-23.92 = -24.52 + 0.60)


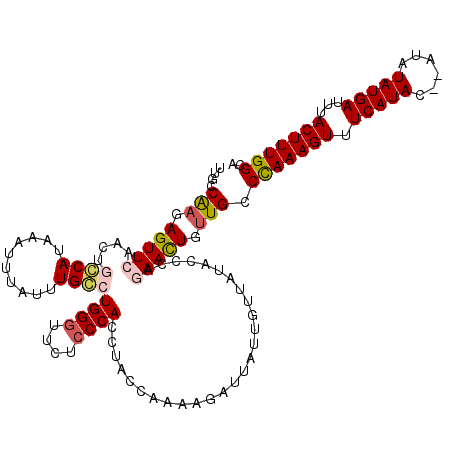
| Location | 12,953,375 – 12,953,493 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.10 |
| Mean single sequence MFE | -25.92 |
| Consensus MFE | -20.76 |
| Energy contribution | -21.28 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.962083 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12953375 118 - 23771897 UUGCCAAGAGUUCAACUGCCAUAAAUUUAUUUGCCUGGUUUCUCCCACCUACCAAAAGAUUAUUGUUAUACCCAGAACUGUUGCCCAAAGUUUCAUAC--AUAUAUGAUUUACUUUGGCA ....(((.(((((..........(((((.((((...(((.......)))...)))))))))..((.......))))))).))).(((((((.(((((.--...)))))...))))))).. ( -21.40) >DroSec_CAF1 70582 118 - 1 UUGCCAAGAGUUCAACUGGCAUAAAUUUAUUUGCCUGGGUUCUCCCACCUACCAAAAGAUUAUUGUUAUACCCAGAAUUGUUGCCUAAAGUUUCAUAC--ACAUAUGAUUUACUUUGGCA .....((.(((((....((((..........))))((((....))))...........................))))).))(((.(((((.(((((.--...)))))...)))))))). ( -25.20) >DroSim_CAF1 75433 118 - 1 UUGCCAAGAGUUCAACUGGCAUAAAUUUAUUUGCCUGGGUUCUCCCACCUACCAAAAGAUUAUUGUUAUACCCCGAAUUGUUGCCCAAAGUUUCAUAC--AUAUAUGAUUUACUUUGGCA ....(((.(((((....((((..........))))((((....))))...........................))))).))).(((((((.(((((.--...)))))...))))))).. ( -27.00) >DroEre_CAF1 76669 118 - 1 UUGUCGAGAGUUCAACUGCCAUAAAUUUAUUUGGAUGGGUUCUCCCACCUACCAAAAGAUUAUUGUUGUACCCAAAACUGUUGGCCAAAGUUUCAUAC--AUAUAUGAUUUACUUUGGCA ....(((.((((....(((.((((..((.(((((.(((((......)))))))))).))...)))).))).....)))).)))((((((((.(((((.--...)))))...)))))))). ( -31.90) >DroYak_CAF1 74433 120 - 1 UUGUCAAGAGUUCAACUGCCAUAAAUUUAUUUGGAUGGGUUCUCCCACAUACCAAAAGAUUAUUGUUGUACCCAAAACUGAUGCCCAAAGUUUCAUACACAUAUAUGAUUUACUUUGGCA .......(.((.((((.......(((((.(((((.((((....))))....))))))))))...)))).)).)........((((.(((((.(((((......)))))...))))))))) ( -24.10) >consensus UUGCCAAGAGUUCAACUGCCAUAAAUUUAUUUGCCUGGGUUCUCCCACCUACCAAAAGAUUAUUGUUAUACCCAGAACUGUUGCCCAAAGUUUCAUAC__AUAUAUGAUUUACUUUGGCA ....(((.(((((....((((..........))))((((....))))...........................))))).))).(((((((.(((((......)))))...))))))).. (-20.76 = -21.28 + 0.52)

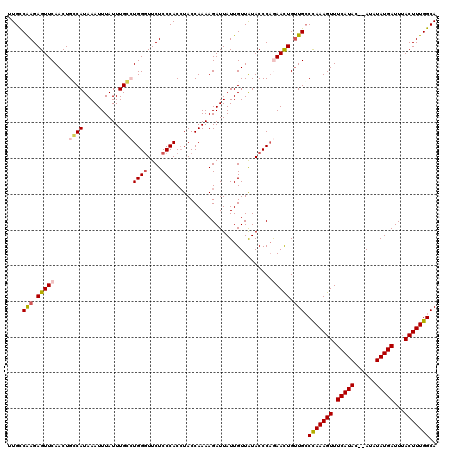
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:23:28 2006