
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,952,560 – 12,952,758 |
| Length | 198 |
| Max. P | 0.968590 |

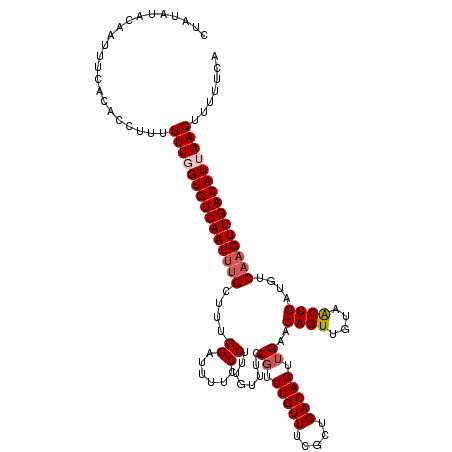
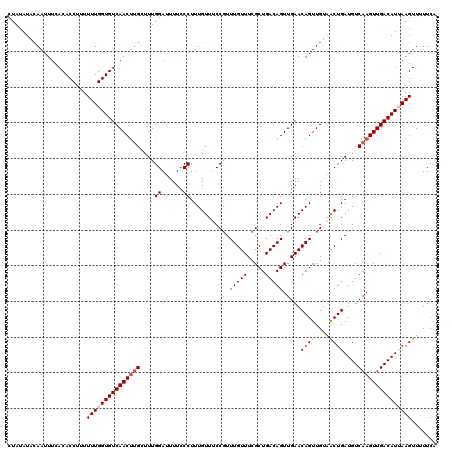
| Location | 12,952,560 – 12,952,680 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.08 |
| Mean single sequence MFE | -26.20 |
| Consensus MFE | -20.76 |
| Energy contribution | -21.60 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.853394 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12952560 120 + 23771897 CUAUAUACAAUUUCACACCUUUUUUGGUGUCAACUUGCUUUGGAUUUUGCCUUUGUUUCCGUUUGUUUCGCUGACAGUUGAACAGUUGUAACUGAUGUCAAGUUGACAUUAAGUUUUUCA ......................((((((((((((((((.(..(...((((..((((((....(((((.....)))))..))))))..)))))..).).)))))))))))))))....... ( -28.20) >DroSec_CAF1 69771 120 + 1 CUAUAUACAAUUUCACACCUUUUUUGGUGUCAACUUGCUUUGGAUUUUCCCUUUGUUUCCGUUUGUUUCGCUGACAGUUGAACAGUUGAAACUGAUGUCAAGUUGACAUUAAGUUUUUCA ......................((((((((((((((((...((.....))....(((((...((((((.((.....)).))))))..)))))....).)))))))))))))))....... ( -27.80) >DroSim_CAF1 74623 120 + 1 CUAUAUACAAUUUCACACCUUUUUUGGUGUCAACUUGCUUUGGAUUUUCCCUUUGUUUCCGUUUGUUUCGCUGACAGUUGAACAGUUGUAACUGAUGUCAAGUUGACAUUAAGUUUUUCA .....(((((((...((((......))))(((((.......((.....))...((((..((.......))..)))))))))..)))))))..(((((((.....)))))))......... ( -25.00) >DroEre_CAF1 75865 120 + 1 CUAUAUAGAAUUUCACACCUUUUUUGGUGUCAACUUGAUUUGGAAUUUCCCUUUGUUGCCCUUUGUUUCGCUGACAGUUGAACAGUUGUUACUGAUGUCAAGUUGACAUAAAGUUUUUCA .......(((...(..(((......)))(((((((((((..((.....))....((.((...((((((.((.....)).))))))..)).))....))))))))))).....)...))). ( -25.90) >DroYak_CAF1 73642 119 + 1 CUAUAUACAAUUUCACACCUUUUUUGGUGUCAACCCGAUUUGGAAUUUCCCUUUGUU-CCGUUUGUUUCGCUGACAGUUGAACAGUUGUUGCUGAUGUCAAGUUGACAUAAAGUUUUUCA ...............((((......))))(((((.......(((((........)))-))...((((.....))))))))).((((....))))(((((.....)))))........... ( -24.10) >consensus CUAUAUACAAUUUCACACCUUUUUUGGUGUCAACUUGCUUUGGAUUUUCCCUUUGUUUCCGUUUGUUUCGCUGACAGUUGAACAGUUGUAACUGAUGUCAAGUUGACAUUAAGUUUUUCA ......................(((((((((((((((....((......))........((.(((((.....))))).))..((((....))))....)))))))))))))))....... (-20.76 = -21.60 + 0.84)

| Location | 12,952,560 – 12,952,680 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.08 |
| Mean single sequence MFE | -23.66 |
| Consensus MFE | -19.66 |
| Energy contribution | -19.66 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968590 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12952560 120 - 23771897 UGAAAAACUUAAUGUCAACUUGACAUCAGUUACAACUGUUCAACUGUCAGCGAAACAAACGGAAACAAAGGCAAAAUCCAAAGCAAGUUGACACCAAAAAAGGUGUGAAAUUGUAUAUAG ............((((...((((((.((((....))))......))))))..........(....)...)))).........((((.((.(((((......))))).)).))))...... ( -23.20) >DroSec_CAF1 69771 120 - 1 UGAAAAACUUAAUGUCAACUUGACAUCAGUUUCAACUGUUCAACUGUCAGCGAAACAAACGGAAACAAAGGGAAAAUCCAAAGCAAGUUGACACCAAAAAAGGUGUGAAAUUGUAUAUAG ...........(((((.....)))))((((((((.....((((((....((.........(....)....((.....))...)).))))))((((......))))))))))))....... ( -26.70) >DroSim_CAF1 74623 120 - 1 UGAAAAACUUAAUGUCAACUUGACAUCAGUUACAACUGUUCAACUGUCAGCGAAACAAACGGAAACAAAGGGAAAAUCCAAAGCAAGUUGACACCAAAAAAGGUGUGAAAUUGUAUAUAG .....((((..(((((.....))))).))))((((...((((...((((((.........(....)....((.....)).......))))))(((......))).)))).))))...... ( -23.80) >DroEre_CAF1 75865 120 - 1 UGAAAAACUUUAUGUCAACUUGACAUCAGUAACAACUGUUCAACUGUCAGCGAAACAAAGGGCAACAAAGGGAAAUUCCAAAUCAAGUUGACACCAAAAAAGGUGUGAAAUUCUAUAUAG ........(((((((((((((((...(((((((....)))..))))..............(....)....((.....))...))))))))))(((......))))))))........... ( -22.30) >DroYak_CAF1 73642 119 - 1 UGAAAAACUUUAUGUCAACUUGACAUCAGCAACAACUGUUCAACUGUCAGCGAAACAAACGG-AACAAAGGGAAAUUCCAAAUCGGGUUGACACCAAAAAAGGUGUGAAAUUGUAUAUAG ........(((((((((((((((.............(((((...(((.......)))....)-))))...((.....))...))))))))))(((......))))))))........... ( -22.30) >consensus UGAAAAACUUAAUGUCAACUUGACAUCAGUUACAACUGUUCAACUGUCAGCGAAACAAACGGAAACAAAGGGAAAAUCCAAAGCAAGUUGACACCAAAAAAGGUGUGAAAUUGUAUAUAG ...........(((((.....)))))(((......)))((((...((((((.........(....)...((......)).......))))))(((......))).))))........... (-19.66 = -19.66 + 0.00)

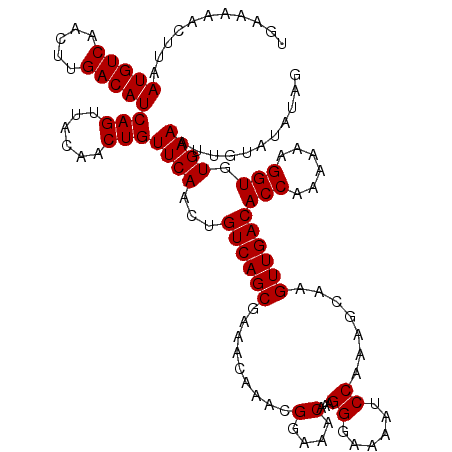

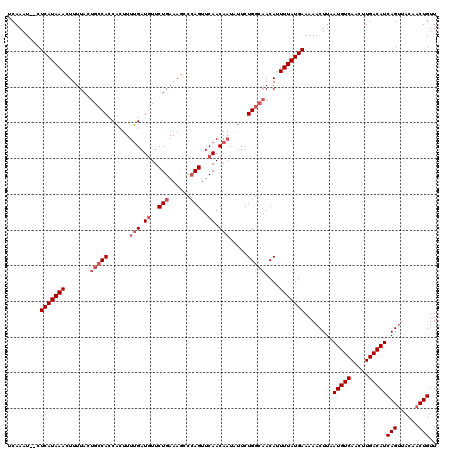
| Location | 12,952,640 – 12,952,758 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.42 |
| Mean single sequence MFE | -19.83 |
| Consensus MFE | -14.24 |
| Energy contribution | -15.64 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.652635 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12952640 118 - 23771897 UCAAAU--CUCAUAAACUUUUACUGCCACCACUUUUGAUGUUCUGAAAGGCCAGUUCAACACUAUUCUGGCAACAUUUUAUGAAAAACUUAAUGUCAACUUGACAUCAGUUACAACUGUU ......--.(((((((((((((..((...((....))..))..))))))(((((............))))).....)))))))..((((..(((((.....))))).))))......... ( -21.10) >DroSec_CAF1 69851 118 - 1 UCAAAU--CUCAUAAACUUUUAUUGCCACCACUUUUGAUGUUCUGAAAGCCAAGUUCAACAAUAUUCUGGCAACAUUUUAUGAAAAACUUAAUGUCAACUUGACAUCAGUUUCAACUGUU ......--.(((((((......((((((......(((.((..((........))..)).))).....))))))...))))))).(((((..(((((.....))))).)))))........ ( -20.20) >DroSim_CAF1 74703 120 - 1 UCAAUUCUCUCAUAAACUUUUACUGCCACCACUUUUGAUGUUCUGAAAACCCAGUUCAACAAUAUUCUGGCAACAUUUUAUGAAAAACUUAAUGUCAACUUGACAUCAGUUACAACUGUU .........(((((((.......(((((......(((.((..(((......)))..)).))).....)))))....)))))))..((((..(((((.....))))).))))......... ( -21.20) >DroEre_CAF1 75945 118 - 1 UCAAAU--CUCAUAAACUUUUACUGCCACCCAUUUGGAUGUUCUGAAAGCCCAGUUCAACAAUUUUCUGUGAACAUUUUAUGAAAAACUUUAUGUCAACUUGACAUCAGUAACAACUGUU ......--.(((((((((((((..((...((....))..))..))))))....(((((.((......)))))))..)))))))........(((((.....)))))((((....)))).. ( -20.80) >DroYak_CAF1 73721 118 - 1 UCAAAU--CUCAUAAACUUUUCCUGCCACCACUUUUGAUUUUCUGAAAGCCCAGUUCAACAAUUUUCUGGCUACAUUUUAUGAAAAACUUUAUGUCAACUUGACAUCAGCAACAACUGUU ......--.(((((((........((((......((((....(((......))).))))........)))).....)))))))........(((((.....)))))(((......))).. ( -15.86) >consensus UCAAAU__CUCAUAAACUUUUACUGCCACCACUUUUGAUGUUCUGAAAGCCCAGUUCAACAAUAUUCUGGCAACAUUUUAUGAAAAACUUAAUGUCAACUUGACAUCAGUUACAACUGUU .........(((((((.......(((((......(((.((..(((......)))..)).))).....)))))....)))))))........(((((.....)))))(((......))).. (-14.24 = -15.64 + 1.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:23:20 2006