
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,948,618 – 12,948,763 |
| Length | 145 |
| Max. P | 0.951114 |

| Location | 12,948,618 – 12,948,738 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.50 |
| Mean single sequence MFE | -31.14 |
| Consensus MFE | -28.32 |
| Energy contribution | -28.16 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951114 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12948618 120 + 23771897 GGGCCACUUUAUUGUUACCAGUUUGUUGCAUAGGUAGGCAAUAAUUUACAGAAAUUAUACAUGACCACUUAAGUUGUCAUUAACGUUAACGACCACCAAACCGAAAGGGAGCGGUCGAAA ..(((((((...(((.((......)).))).)))).))).(((((((....)))))))..(((((.((....)).))))).........(((((.(....((....))..).)))))... ( -29.30) >DroSec_CAF1 65746 120 + 1 GGGCCACUUUAUUGUUACCAGUUUGUUGCAUAGGUGGGCAAUAAUUUACAGAAAUUAUACAUGACCACUUAAGUUGUCAUUAACGUUAACGACCACCAAACCGAAAGGGAGCGGUCGAAA .(.((((((...(((.((......)).))).)))))).).(((((((....)))))))..(((((.((....)).))))).........(((((.(....((....))..).)))))... ( -31.80) >DroSim_CAF1 70545 120 + 1 GGGCCACUUUAUUGUUACCAGUUUGUUGCAUAGGUAGGCAAUAAUUUACAGAAAUUAUACAUGACCACUUAAGUUGUCAUUAACGUUAACGACCACCAAACCGAAAGGGAGCGGUCGAAA ..(((((((...(((.((......)).))).)))).))).(((((((....)))))))..(((((.((....)).))))).........(((((.(....((....))..).)))))... ( -29.30) >DroEre_CAF1 71877 120 + 1 GGGCCACUUUAUUGUUACCAGUUUGUUGCAUAGGUAGGCAAUAAUUUACAGAAAUUAUACAUGACCACUUAAGUUGUCGUUAACGUUAACGACCGCCAAACCAAAAGGGAGCGGUCGAAA ..(((((((...(((.((......)).))).)))).))).(((((((....)))))))..(((((.((....)).))))).........(((((((....((....))..)))))))... ( -30.60) >DroYak_CAF1 69623 120 + 1 GGGCCACUUUAUUGUUACCAGUUUGUUGCAUAGGUAGGCAAUAAUUUACAGAAAUUAUACAUGACCACUUAAGUUGUCAUUAACGUUAACGACCGCCAAACCGAAAGGGAGCGGUCGAAA ..(((((((...(((.((......)).))).)))).))).(((((((....)))))))..(((((.((....)).))))).........(((((((....((....))..)))))))... ( -34.70) >consensus GGGCCACUUUAUUGUUACCAGUUUGUUGCAUAGGUAGGCAAUAAUUUACAGAAAUUAUACAUGACCACUUAAGUUGUCAUUAACGUUAACGACCACCAAACCGAAAGGGAGCGGUCGAAA ..(((((((...(((.((......)).))).)))).))).(((((((....)))))))..(((((.((....)).))))).........(((((.(....((....))..).)))))... (-28.32 = -28.16 + -0.16)


| Location | 12,948,658 – 12,948,763 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.23 |
| Mean single sequence MFE | -32.86 |
| Consensus MFE | -22.07 |
| Energy contribution | -21.75 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.677386 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12948658 105 + 23771897 AUAAUUUACAGAAAUUAUACAUGACCACUUAAGUUGUCAUUAACGUUAACGACCACCAAACCGAAAGGGAGCGGUCGAAAUGUGUGUCGGGCCAUUAUG------------GCCUCG--- (((((((....)))))))....((((((....((((....)))).....(((((.(....((....))..).)))))....))).)))(((((.....)------------))))..--- ( -31.10) >DroSec_CAF1 65786 105 + 1 AUAAUUUACAGAAAUUAUACAUGACCACUUAAGUUGUCAUUAACGUUAACGACCACCAAACCGAAAGGGAGCGGUCGAAAUGUGUGUCGGGCCAUUAUG------------GCCUCG--- (((((((....)))))))....((((((....((((....)))).....(((((.(....((....))..).)))))....))).)))(((((.....)------------))))..--- ( -31.10) >DroSim_CAF1 70585 105 + 1 AUAAUUUACAGAAAUUAUACAUGACCACUUAAGUUGUCAUUAACGUUAACGACCACCAAACCGAAAGGGAGCGGUCGAAAUGUGUGUCGGGCCAUUAUG------------GCCUCG--- (((((((....)))))))....((((((....((((....)))).....(((((.(....((....))..).)))))....))).)))(((((.....)------------))))..--- ( -31.10) >DroEre_CAF1 71917 117 + 1 AUAAUUUACAGAAAUUAUACAUGACCACUUAAGUUGUCGUUAACGUUAACGACCGCCAAACCAAAAGGGAGCGGUCGAAAUGUAUGUCGGGGCAUUAUGAAAUGUUUAAGAGCCCCG--- ...............((((((((((.((....)).)))...........(((((((....((....))..)))))))..))))))).((((((.(((.........)))..))))))--- ( -34.40) >DroYak_CAF1 69663 120 + 1 AUAAUUUACAGAAAUUAUACAUGACCACUUAAGUUGUCAUUAACGUUAACGACCGCCAAACCGAAAGGGAGCGGUCGAAAUGUGUGUCGGGGCAUUAUGAAAUGUUUAAGAGCCUCGCAG (((((((....)))))))..(((((.((....)).)))))..(((((..(((((((....((....))..))))))).))))).((.((((((.(((.........)))..)))))))). ( -36.60) >consensus AUAAUUUACAGAAAUUAUACAUGACCACUUAAGUUGUCAUUAACGUUAACGACCACCAAACCGAAAGGGAGCGGUCGAAAUGUGUGUCGGGCCAUUAUG____________GCCUCG___ ...............((((((((((.((....)).)))...........(((((.(....((....))..).)))))..))))))).((((......................))))... (-22.07 = -21.75 + -0.32)


| Location | 12,948,658 – 12,948,763 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.23 |
| Mean single sequence MFE | -29.23 |
| Consensus MFE | -21.42 |
| Energy contribution | -21.02 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.663405 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
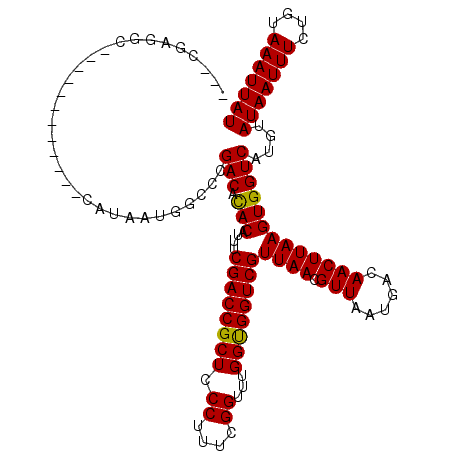
>3L_DroMel_CAF1 12948658 105 - 23771897 ---CGAGGC------------CAUAAUGGCCCGACACACAUUUCGACCGCUCCCUUUCGGUUUGGUGGUCGUUAACGUUAAUGACAACUUAAGUGGUCAUGUAUAAUUUCUGUAAAUUAU ---.(.(((------------(.....)))))(((.(((....((((((((.((....))...))))))))((((.(((......)))))))))))))....(((((((....))))))) ( -28.30) >DroSec_CAF1 65786 105 - 1 ---CGAGGC------------CAUAAUGGCCCGACACACAUUUCGACCGCUCCCUUUCGGUUUGGUGGUCGUUAACGUUAAUGACAACUUAAGUGGUCAUGUAUAAUUUCUGUAAAUUAU ---.(.(((------------(.....)))))(((.(((....((((((((.((....))...))))))))((((.(((......)))))))))))))....(((((((....))))))) ( -28.30) >DroSim_CAF1 70585 105 - 1 ---CGAGGC------------CAUAAUGGCCCGACACACAUUUCGACCGCUCCCUUUCGGUUUGGUGGUCGUUAACGUUAAUGACAACUUAAGUGGUCAUGUAUAAUUUCUGUAAAUUAU ---.(.(((------------(.....)))))(((.(((....((((((((.((....))...))))))))((((.(((......)))))))))))))....(((((((....))))))) ( -28.30) >DroEre_CAF1 71917 117 - 1 ---CGGGGCUCUUAAACAUUUCAUAAUGCCCCGACAUACAUUUCGACCGCUCCCUUUUGGUUUGGCGGUCGUUAACGUUAACGACAACUUAAGUGGUCAUGUAUAAUUUCUGUAAAUUAU ---((((((..................))))))..((((((...(((((((.((....))...((..(((((((....)))))))..))..)))))))))))))................ ( -34.77) >DroYak_CAF1 69663 120 - 1 CUGCGAGGCUCUUAAACAUUUCAUAAUGCCCCGACACACAUUUCGACCGCUCCCUUUCGGUUUGGCGGUCGUUAACGUUAAUGACAACUUAAGUGGUCAUGUAUAAUUUCUGUAAAUUAU .((((.(((..................))).)(((.(((....((((((((.((....))...))))))))((((.(((......))))))))))))).............)))...... ( -26.47) >consensus ___CGAGGC____________CAUAAUGGCCCGACACACAUUUCGACCGCUCCCUUUCGGUUUGGUGGUCGUUAACGUUAAUGACAACUUAAGUGGUCAUGUAUAAUUUCUGUAAAUUAU ................................(((.(((....((((((((.((....))...))))))))((((.(((......)))))))))))))....(((((((....))))))) (-21.42 = -21.02 + -0.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:23:04 2006