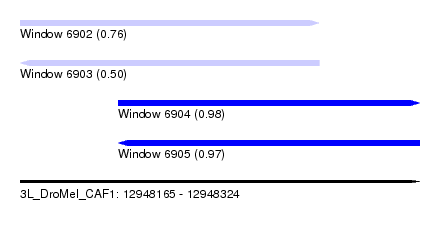
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,948,165 – 12,948,324 |
| Length | 159 |
| Max. P | 0.982560 |

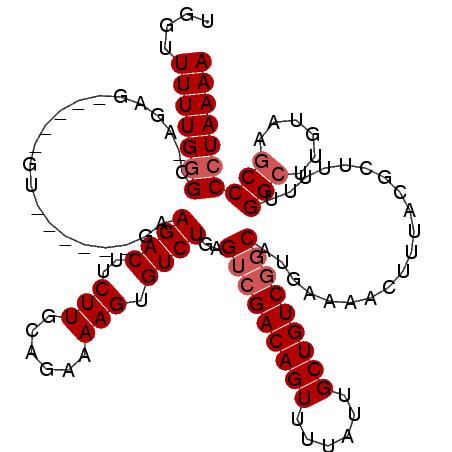
| Location | 12,948,165 – 12,948,284 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.35 |
| Mean single sequence MFE | -33.54 |
| Consensus MFE | -24.10 |
| Energy contribution | -24.90 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.756970 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
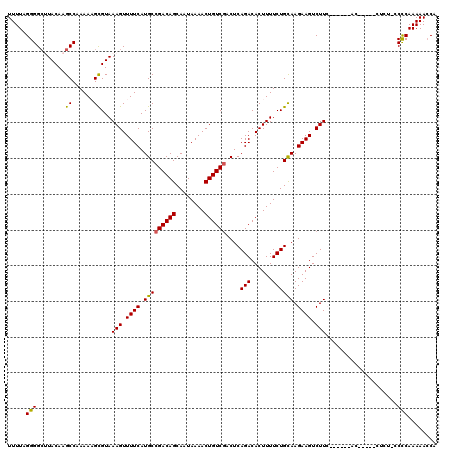
>3L_DroMel_CAF1 12948165 119 + 23771897 UGGUUUUUGGGC-AGAGGUUGAGUUUCGCCGAAGACUUCUUGCAGAAAAGUGUCUGAGACGACAGUUUUAUUGCUGUCGGCAUGAAAACUUUACGCUUUUUGGCUUGUAAGCCCCUAAAA ...(((..((((-(((((((..(......)...))))))).((.(((((((((..(((.(((((((......)))))))...(....)))).))))))))).))......))))..))). ( -35.60) >DroSec_CAF1 65388 105 + 1 UGGUUUUUGGGU-AGAG--------------AAGACUUCUUACAGAAAAGUGUCUGAGUCGACAGUUUUAUUGCUGUCGGCAUGAAAACUUUACGCUUUUUGGCUUGUAUGCCCCUAAAA ....((((((((-((((--------------.((((..(((......))).))))..(((((((((......))))))))).......)))))).......(((......))).)))))) ( -31.90) >DroSim_CAF1 69742 105 + 1 UGGUUUUUGGGC-AGAG--------------AAGACUUCUUGCAGAAAAGUGUCUGAGUCGACAGUUUUAUUGCUGUCGGCAUGAAAACUUUACGCUUUUUGGCUUGUACUCCCCUAAAA ....(((((((.-.(((--------------(((....)))((.(((((((((....(((((((((......))))))))).(....)....))))))))).)).....))).))))))) ( -31.70) >DroEre_CAF1 71466 113 + 1 UGGUUUUUGAGG--G-----AGGUGCUCCAGAAGACUUCUUGCAGAAAAGUGUCUGAGUAGACAGUUUUAUUGCUGUCGGCAUGAAAACUUUACGCAUUUUGGCUUGUAAGCCCCUAAAA .........(((--(-----((((((....((((..(((.((((((......))).....((((((......)))))).))).)))..))))..)))))))((((....))))))).... ( -29.10) >DroYak_CAF1 69183 120 + 1 CGCUUUUUGGGGUGGAGGUUAAGUGCCCAAGAAGACUUCUUGCAGAAAAGUGUCUGAGUCGACAGUUUUAUUGCUGUCGGCAUGAAAACUUUACACAUUUUGGCUUGUAAGCCCCUAAAA ....((((((((((((((((..(((((......((((.(..(((......)))..)))))((((((......)))))))))))...)))))).))).....((((....))))))))))) ( -39.40) >consensus UGGUUUUUGGGC_AGAG_____GU______GAAGACUUCUUGCAGAAAAGUGUCUGAGUCGACAGUUUUAUUGCUGUCGGCAUGAAAACUUUACGCUUUUUGGCUUGUAAGCCCCUAAAA ....(((((((.....................((((..(((......))).))))..(((((((((......)))))))))....................(((......)))))))))) (-24.10 = -24.90 + 0.80)

| Location | 12,948,165 – 12,948,284 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.35 |
| Mean single sequence MFE | -24.72 |
| Consensus MFE | -18.34 |
| Energy contribution | -18.06 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.74 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12948165 119 - 23771897 UUUUAGGGGCUUACAAGCCAAAAAGCGUAAAGUUUUCAUGCCGACAGCAAUAAAACUGUCGUCUCAGACACUUUUCUGCAAGAAGUCUUCGGCGAAACUCAACCUCU-GCCCAAAAACCA ......((((......((......))((..((((((...(((((..(((..((((.((((......)))).)))).))).((....)))))))))))))..))....-))))........ ( -27.70) >DroSec_CAF1 65388 105 - 1 UUUUAGGGGCAUACAAGCCAAAAAGCGUAAAGUUUUCAUGCCGACAGCAAUAAAACUGUCGACUCAGACACUUUUCUGUAAGAAGUCUU--------------CUCU-ACCCAAAAACCA ((((.(((((......))).......(..(((.((((....((((((........))))))...((((......))))...)))).)))--------------..).-.)).)))).... ( -21.00) >DroSim_CAF1 69742 105 - 1 UUUUAGGGGAGUACAAGCCAAAAAGCGUAAAGUUUUCAUGCCGACAGCAAUAAAACUGUCGACUCAGACACUUUUCUGCAAGAAGUCUU--------------CUCU-GCCCAAAAACCA .....(((((((....((...(((((.....)))))...))((((((........))))))))))(((.((((((.....))))))..)--------------))..-.)))........ ( -23.50) >DroEre_CAF1 71466 113 - 1 UUUUAGGGGCUUACAAGCCAAAAUGCGUAAAGUUUUCAUGCCGACAGCAAUAAAACUGUCUACUCAGACACUUUUCUGCAAGAAGUCUUCUGGAGCACCU-----C--CCUCAAAAACCA (((((((((((....))))....((((((.((((((..(((.....)))..))))))...)))(((((.((((((.....))))))..))))).)))...-----.--))).)))).... ( -24.50) >DroYak_CAF1 69183 120 - 1 UUUUAGGGGCUUACAAGCCAAAAUGUGUAAAGUUUUCAUGCCGACAGCAAUAAAACUGUCGACUCAGACACUUUUCUGCAAGAAGUCUUCUUGGGCACUUAACCUCCACCCCAAAAAGCG .....((((.(((((..........))))).(((....(((((((((........)))))....((((......))))(((((.....)))))))))...))).....))))........ ( -26.90) >consensus UUUUAGGGGCUUACAAGCCAAAAAGCGUAAAGUUUUCAUGCCGACAGCAAUAAAACUGUCGACUCAGACACUUUUCUGCAAGAAGUCUUC______AC_____CUCU_CCCCAAAAACCA .....(((........((......))...(((.((((.(((((((((........))))))....(((......)))))).)))).)))....................)))........ (-18.34 = -18.06 + -0.28)

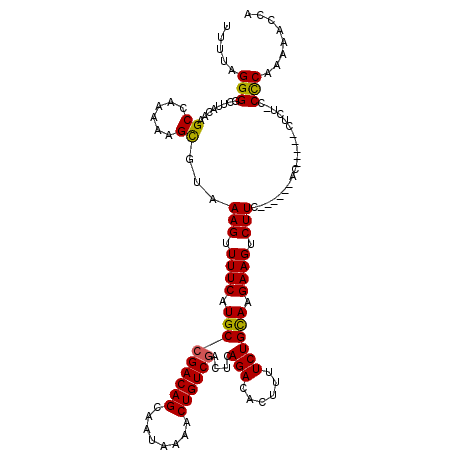
| Location | 12,948,204 – 12,948,324 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.92 |
| Mean single sequence MFE | -31.54 |
| Consensus MFE | -28.94 |
| Energy contribution | -29.82 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.92 |
| SVM RNA-class probability | 0.982560 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
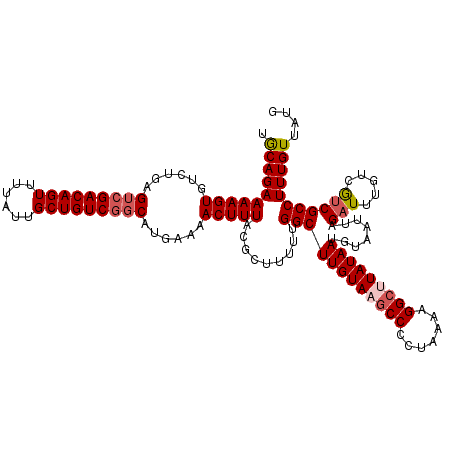
>3L_DroMel_CAF1 12948204 120 + 23771897 UGCAGAAAAGUGUCUGAGACGACAGUUUUAUUGCUGUCGGCAUGAAAACUUUACGCUUUUUGGCUUGUAAGCCCCUAAAAGGCUUAUAAUGUAAUUAGAUUUGUCGUCGCCUUUGUUAUG .((((((((((((..(((.(((((((......)))))))...(....)))).)))))))))((((((((((((.......)))))))))........(((.....))))))..))).... ( -33.70) >DroSec_CAF1 65413 120 + 1 UACAGAAAAGUGUCUGAGUCGACAGUUUUAUUGCUGUCGGCAUGAAAACUUUACGCUUUUUGGCUUGUAUGCCCCUAAAAGGCUUAUAAUGUAAUUAGAUUUGUCGUCGCCUUUGUUAUG ....(((((((((....(((((((((......))))))))).(....)....)))))))))((((((((.(((.......))).)))))........(((.....))))))......... ( -31.00) >DroSim_CAF1 69767 120 + 1 UGCAGAAAAGUGUCUGAGUCGACAGUUUUAUUGCUGUCGGCAUGAAAACUUUACGCUUUUUGGCUUGUACUCCCCUAAAAGGCUUAUAAUGUAAUUAGAUUUGUCGUCGCCUUUGUUAUG .((.(((((((((....(((((((((......))))))))).(....)....))))))))).)).............((((((...((((...))))(((.....)))))))))...... ( -29.40) >DroEre_CAF1 71499 120 + 1 UGCAGAAAAGUGUCUGAGUAGACAGUUUUAUUGCUGUCGGCAUGAAAACUUUACGCAUUUUGGCUUGUAAGCCCCUAAAAGGCUUAUAAUGUAAUUAGAUUUGUCACCGCCUUUGUUUUG ..((((......))))....((((((......))))))(((.((((((((((((((......))(((((((((.......))))))))).))))..)).))).)))..)))......... ( -29.20) >DroYak_CAF1 69223 120 + 1 UGCAGAAAAGUGUCUGAGUCGACAGUUUUAUUGCUGUCGGCAUGAAAACUUUACACAUUUUGGCUUGUAAGCCCCUAAAAGGCUUAUAAUGUAAUUAGAUUUGUCGUCGCCUUUGUUAUU .((((((((((((.((((((((((((......))))))))).(....)..))).)))))))((((((((((((.......)))))))))........(((.....))))))))))).... ( -34.40) >consensus UGCAGAAAAGUGUCUGAGUCGACAGUUUUAUUGCUGUCGGCAUGAAAACUUUACGCUUUUUGGCUUGUAAGCCCCUAAAAGGCUUAUAAUGUAAUUAGAUUUGUCGUCGCCUUUGUUAUG .((((((((((......(((((((((......)))))))))......))))).........((((((((((((.......)))))))))........(((.....))))))))))).... (-28.94 = -29.82 + 0.88)

| Location | 12,948,204 – 12,948,324 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.92 |
| Mean single sequence MFE | -25.52 |
| Consensus MFE | -22.12 |
| Energy contribution | -22.60 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.969891 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
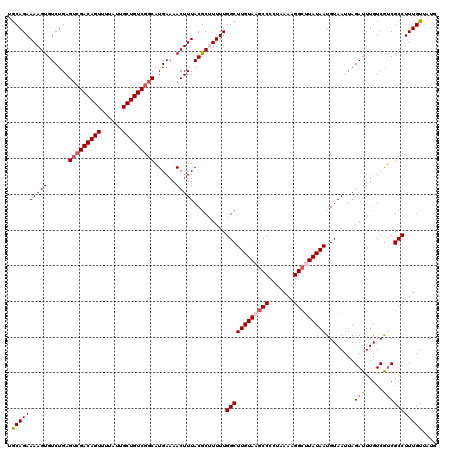
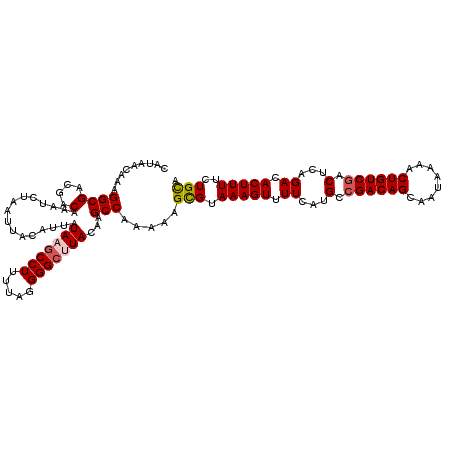
>3L_DroMel_CAF1 12948204 120 - 23771897 CAUAACAAAGGCGACGACAAAUCUAAUUACAUUAUAAGCCUUUUAGGGGCUUACAAGCCAAAAAGCGUAAAGUUUUCAUGCCGACAGCAAUAAAACUGUCGUCUCAGACACUUUUCUGCA ..........((((((((........((((....(((((((.....)))))))...((......))))))((((((..(((.....)))..))))))))))))..(((......))))). ( -29.20) >DroSec_CAF1 65413 120 - 1 CAUAACAAAGGCGACGACAAAUCUAAUUACAUUAUAAGCCUUUUAGGGGCAUACAAGCCAAAAAGCGUAAAGUUUUCAUGCCGACAGCAAUAAAACUGUCGACUCAGACACUUUUCUGUA .........(((((.(((...................((((.....))))......((......)).....))).))..)))(((((........)))))....((((......)))).. ( -22.30) >DroSim_CAF1 69767 120 - 1 CAUAACAAAGGCGACGACAAAUCUAAUUACAUUAUAAGCCUUUUAGGGGAGUACAAGCCAAAAAGCGUAAAGUUUUCAUGCCGACAGCAAUAAAACUGUCGACUCAGACACUUUUCUGCA ......((((((.........................))))))(((..((((....((...(((((.....)))))...))((((((........))))))........))))..))).. ( -24.81) >DroEre_CAF1 71499 120 - 1 CAAAACAAAGGCGGUGACAAAUCUAAUUACAUUAUAAGCCUUUUAGGGGCUUACAAGCCAAAAUGCGUAAAGUUUUCAUGCCGACAGCAAUAAAACUGUCUACUCAGACACUUUUCUGCA .........((((....)........((((....(((((((.....)))))))...((......)))))).........)))....(((..((((.(((((....))))).)))).))). ( -27.20) >DroYak_CAF1 69223 120 - 1 AAUAACAAAGGCGACGACAAAUCUAAUUACAUUAUAAGCCUUUUAGGGGCUUACAAGCCAAAAUGUGUAAAGUUUUCAUGCCGACAGCAAUAAAACUGUCGACUCAGACACUUUUCUGCA .........(((...((....))...........(((((((.....)))))))...)))....(((..(((((.((...(.((((((........)))))).)...)).)))))...))) ( -24.10) >consensus CAUAACAAAGGCGACGACAAAUCUAAUUACAUUAUAAGCCUUUUAGGGGCUUACAAGCCAAAAAGCGUAAAGUUUUCAUGCCGACAGCAAUAAAACUGUCGACUCAGACACUUUUCUGCA .........((((....)................(((((((.....)))))))...))).....(((.(((((.((...(.((((((........)))))).)...)).)))))..))). (-22.12 = -22.60 + 0.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:23:01 2006