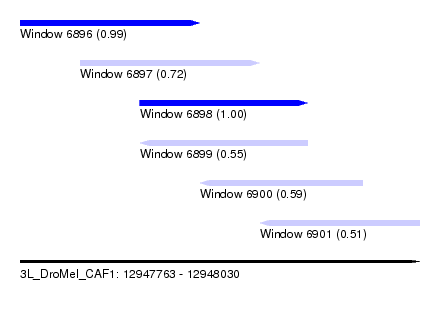
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,947,763 – 12,948,030 |
| Length | 267 |
| Max. P | 0.999823 |

| Location | 12,947,763 – 12,947,883 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.33 |
| Mean single sequence MFE | -43.20 |
| Consensus MFE | -37.46 |
| Energy contribution | -37.82 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.38 |
| SVM RNA-class probability | 0.993239 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12947763 120 + 23771897 GGGAUCCAGGCGCGAAAGCCACUGACACAGCUGGAACUCGGCACCAAACAGUUUCGGAUUCCCAUGCCAUCAGCGAACUCAGCGGAGCCGCAAAUCUUUGCGGUCUCGUCUUAAUGGCAA (((((((.(((......)))((((.....((((.....))))......))))...)))))))..((((((..((.......)).(((((((((....)))))).)))......)))))). ( -44.30) >DroSec_CAF1 64990 120 + 1 GGGAUCCGGGCGCGAAAGCCACUGACACAGCUGGAACUCGGCACCAAACAGUUUCGGAUUCCCAUGCCAUCAGCGAACCCAGCGCAGCCGCAAAUCUUUGCGGUCUCGUCUUAAUGGCAA (((((((((((......)))((((.....((((.....))))......))))..))))))))..((((((..(((.......))).(((((((....))))))).........)))))). ( -44.00) >DroSim_CAF1 69342 120 + 1 GGGAUCCGGGCGCGAAAGCCACUGACACAGCUGGAACUCGGCACCAAACAGUUUCGGAUUCCCAUGCCAUCAGCGAACCCAGCGGAGCCGCAAAUCUUUGCGGUCUCGUCUUAAUGGCAA (((((((((((......)))((((.....((((.....))))......))))..))))))))..((((((..((.......)).(((((((((....)))))).)))......)))))). ( -45.90) >DroEre_CAF1 71041 116 + 1 GGGAUCCGGGCGCGAAAGCCA----CACAGCUGGAACUCGGCACCAAACAGUUUCAGAUUGCCAUGCCAUCAGCAAACUCAGCGGAGCCGCAAAUCUUUGCGGUCUCGUCUUAAUGGCAA ........(((((....))..----.....((((((((.(........)))))))))...))).((((((..((.......)).(((((((((....)))))).)))......)))))). ( -39.60) >DroYak_CAF1 68758 120 + 1 GGGAUCCGGGCGCGAAAGCCACUGACACAGCUGGAACUCGGCACCAAACAGUUUCAGAUUGCCAGGCCAUCAGCAAACUCAGCGGAGCCGCAAAUCUUUGCGGUCUCCUCUUAAUGGCAA ((....(((..((....))..)))...(((((((((((.(........))))))))).)))))..(((((..((.......))((((((((((....)))))).)))).....))))).. ( -42.20) >consensus GGGAUCCGGGCGCGAAAGCCACUGACACAGCUGGAACUCGGCACCAAACAGUUUCGGAUUCCCAUGCCAUCAGCGAACUCAGCGGAGCCGCAAAUCUUUGCGGUCUCGUCUUAAUGGCAA ((((....(((......)))..........((((((((.(........)))))))))..)))).((((((..((.......))(..(((((((....)))))))..)......)))))). (-37.46 = -37.82 + 0.36)



| Location | 12,947,803 – 12,947,923 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.00 |
| Mean single sequence MFE | -34.66 |
| Consensus MFE | -28.44 |
| Energy contribution | -29.04 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.720819 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12947803 120 + 23771897 GCACCAAACAGUUUCGGAUUCCCAUGCCAUCAGCGAACUCAGCGGAGCCGCAAAUCUUUGCGGUCUCGUCUUAAUGGCAAAGAAAUUGUGAUUUAUGUCUCGAGAUUUGUACAUUUGGGC ((.((((((((((((((....)).((((((..((.......)).(((((((((....)))))).)))......))))))..))))))))(((....)))...............)))))) ( -36.90) >DroSec_CAF1 65030 120 + 1 GCACCAAACAGUUUCGGAUUCCCAUGCCAUCAGCGAACCCAGCGCAGCCGCAAAUCUUUGCGGUCUCGUCUUAAUGGCAAAGAAAUUGUGAUUUAUGUCUCGAGAUUUGUACAUUUAGGC ...((..((((((((((....)).((((((..(((.......))).(((((((....))))))).........))))))..))))))))(((....)))..................)). ( -31.40) >DroSim_CAF1 69382 120 + 1 GCACCAAACAGUUUCGGAUUCCCAUGCCAUCAGCGAACCCAGCGGAGCCGCAAAUCUUUGCGGUCUCGUCUUAAUGGCAAAGAAAUUGUGAUUUAUGUCUCGAGAUUUGUACAUUUAGGC ...((..((((((((((....)).((((((..((.......)).(((((((((....)))))).)))......))))))..))))))))(((....)))..................)). ( -33.30) >DroEre_CAF1 71077 120 + 1 GCACCAAACAGUUUCAGAUUGCCAUGCCAUCAGCAAACUCAGCGGAGCCGCAAAUCUUUGCGGUCUCGUCUUAAUGGCAAAGAAAUUGUGAUUUAUGUCUCGAGAUUUGUGCAUUUAGGC ((((...((((((((...(((((((((.....)).......(((..(((((((....)))))))..)))....))))))).))))))))(((....))).........))))........ ( -37.90) >DroYak_CAF1 68798 120 + 1 GCACCAAACAGUUUCAGAUUGCCAGGCCAUCAGCAAACUCAGCGGAGCCGCAAAUCUUUGCGGUCUCCUCUUAAUGGCAAAGAAAUUGUGAUUUAUGUCUCAAGAUUUGUACAUUUAGGC ...((..((((((((...((((((.((.....))......((.((((((((((....)))))).)))).))...)))))).))))))))(((....)))..................)). ( -33.80) >consensus GCACCAAACAGUUUCGGAUUCCCAUGCCAUCAGCGAACUCAGCGGAGCCGCAAAUCUUUGCGGUCUCGUCUUAAUGGCAAAGAAAUUGUGAUUUAUGUCUCGAGAUUUGUACAUUUAGGC ...((..((((((((((....)).((((((..((.......))(..(((((((....)))))))..)......))))))..))))))))(((....)))..................)). (-28.44 = -29.04 + 0.60)

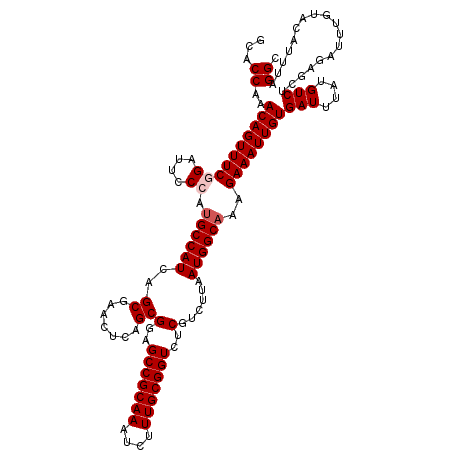
| Location | 12,947,843 – 12,947,955 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.52 |
| Mean single sequence MFE | -34.06 |
| Consensus MFE | -31.34 |
| Energy contribution | -31.82 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.92 |
| SVM decision value | 4.17 |
| SVM RNA-class probability | 0.999823 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12947843 112 + 23771897 AGCGGAGCCGCAAAUCUUUGCGGUCUCGUCUUAAUGGCAAAGAAAUUGUGAUUUAUGUCUCGAGAUUUGUACAUUUGGGCUGCUUGACU--CAAGCGUAUGUAUAUA------UAUACAA ....(((((((((....)))))).)))((((.(((((((((....(((.(((....))).)))..))))).)))).)))).(((((...--)))))((((((....)------))))).. ( -34.90) >DroSec_CAF1 65070 110 + 1 AGCGCAGCCGCAAAUCUUUGCGGUCUCGUCUUAAUGGCAAAGAAAUUGUGAUUUAUGUCUCGAGAUUUGUACAUUUAGGCUGCUUGACU--CAAGCGUAUGUAUA--------UAUACAC ...(..(((((((....)))))))..)((((.(((((((((....(((.(((....))).)))..))))).)))).)))).(((((...--)))))(((((....--------))))).. ( -30.40) >DroSim_CAF1 69422 112 + 1 AGCGGAGCCGCAAAUCUUUGCGGUCUCGUCUUAAUGGCAAAGAAAUUGUGAUUUAUGUCUCGAGAUUUGUACAUUUAGGCUGCUUGACU--CAAGCGAAUGUAUAUA------UAUACAC ....(((((((((....)))))).)))((((.(((((((((....(((.(((....))).)))..))))).)))).))))((((((...--))))))..(((((...------.))))). ( -32.90) >DroEre_CAF1 71117 110 + 1 AGCGGAGCCGCAAAUCUUUGCGGUCUCGUCUUAAUGGCAAAGAAAUUGUGAUUUAUGUCUCGAGAUUUGUGCAUUUAGGCUGCUUGACA--CUAGCGUCUGUA--------UGUAUACAC .(((..(((((((....)))))))..))).......(((((....(((.(((....))).)))..)))))((((.(((((.(((.....--..)))))))).)--------)))...... ( -32.50) >DroYak_CAF1 68838 120 + 1 AGCGGAGCCGCAAAUCUUUGCGGUCUCCUCUUAAUGGCAAAGAAAUUGUGAUUUAUGUCUCAAGAUUUGUACAUUUAGGCUGCUUGACACACAAGCGUAUGUAUAUAUGGAUGUAUACAC (((((((((((((....)))))).)))).((.(((((((((....(((.(((....))).)))..))))).)))).)))))(((((.....)))))...((((((((....)))))))). ( -39.60) >consensus AGCGGAGCCGCAAAUCUUUGCGGUCUCGUCUUAAUGGCAAAGAAAUUGUGAUUUAUGUCUCGAGAUUUGUACAUUUAGGCUGCUUGACU__CAAGCGUAUGUAUAUA______UAUACAC ...(..(((((((....)))))))..)((((.(((((((((....(((.(((....))).)))..))))).)))).)))).(((((.....)))))...((((((........)))))). (-31.34 = -31.82 + 0.48)


| Location | 12,947,843 – 12,947,955 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.52 |
| Mean single sequence MFE | -25.25 |
| Consensus MFE | -20.78 |
| Energy contribution | -21.42 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.547749 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12947843 112 - 23771897 UUGUAUA------UAUAUACAUACGCUUG--AGUCAAGCAGCCCAAAUGUACAAAUCUCGAGACAUAAAUCACAAUUUCUUUGCCAUUAAGACGAGACCGCAAAGAUUUGCGGCUCCGCU .(((((.------...)))))...(((((--...)))))(((.............(((((.((......))......((((.......)))))))))((((((....))))))....))) ( -22.30) >DroSec_CAF1 65070 110 - 1 GUGUAUA--------UAUACAUACGCUUG--AGUCAAGCAGCCUAAAUGUACAAAUCUCGAGACAUAAAUCACAAUUUCUUUGCCAUUAAGACGAGACCGCAAAGAUUUGCGGCUGCGCU ((((((.--------.....))))))...--......((((((............(((((.((......))......((((.......)))))))))..((((....))))))))))... ( -26.90) >DroSim_CAF1 69422 112 - 1 GUGUAUA------UAUAUACAUUCGCUUG--AGUCAAGCAGCCUAAAUGUACAAAUCUCGAGACAUAAAUCACAAUUUCUUUGCCAUUAAGACGAGACCGCAAAGAUUUGCGGCUCCGCU ((((((.------...))))))..(((((--...)))))(((.............(((((.((......))......((((.......)))))))))((((((....))))))....))) ( -23.00) >DroEre_CAF1 71117 110 - 1 GUGUAUACA--------UACAGACGCUAG--UGUCAAGCAGCCUAAAUGCACAAAUCUCGAGACAUAAAUCACAAUUUCUUUGCCAUUAAGACGAGACCGCAAAGAUUUGCGGCUCCGCU ((((((...--------....((((....--)))).((....))..))))))...(((((.((......))......((((.......)))))))))((((((....))))))....... ( -21.80) >DroYak_CAF1 68838 120 - 1 GUGUAUACAUCCAUAUAUACAUACGCUUGUGUGUCAAGCAGCCUAAAUGUACAAAUCUUGAGACAUAAAUCACAAUUUCUUUGCCAUUAAGAGGAGACCGCAAAGAUUUGCGGCUCCGCU (((((((......)))))))....((((((((.(((((..................))))).)))))).........((((.......))))((((.((((((....)))))))))))). ( -32.27) >consensus GUGUAUA______UAUAUACAUACGCUUG__AGUCAAGCAGCCUAAAUGUACAAAUCUCGAGACAUAAAUCACAAUUUCUUUGCCAUUAAGACGAGACCGCAAAGAUUUGCGGCUCCGCU .((((((........))))))...(((((.....)))))(((.............(((((.((......))......((((.......)))))))))((((((....))))))....))) (-20.78 = -21.42 + 0.64)


| Location | 12,947,883 – 12,947,992 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.90 |
| Mean single sequence MFE | -18.62 |
| Consensus MFE | -12.82 |
| Energy contribution | -14.06 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.588624 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12947883 109 - 23771897 ACAAGGAAACUAAAUCGUGUGCAUCAUCUCAAAU---AUAUUGUAUA------UAUAUACAUACGCUUG--AGUCAAGCAGCCCAAAUGUACAAAUCUCGAGACAUAAAUCACAAUUUCU ...((....)).....(((.......((((....---....(((((.------...)))))...(((((--...)))))....................)))).......)))....... ( -16.84) >DroSec_CAF1 65110 98 - 1 ACAAGGAAACUAAAUCGUGUGCAUU------------AAAGUGUAUA--------UAUACAUACGCUUG--AGUCAAGCAGCCUAAAUGUACAAAUCUCGAGACAUAAAUCACAAUUUCU ...((....))...(((((((((((------------...(((((..--------..)))))..(((((--...)))))......)))))))).....)))................... ( -18.10) >DroSim_CAF1 69462 100 - 1 ACAAGGAAACUAAAUCGUGUGCAUU------------AAAGUGUAUA------UAUAUACAUUCGCUUG--AGUCAAGCAGCCUAAAUGUACAAAUCUCGAGACAUAAAUCACAAUUUCU ...((....))...(((((((((((------------..(((((((.------...))))))).(((((--...)))))......)))))))).....)))................... ( -19.40) >DroEre_CAF1 71157 110 - 1 CCAUCAAAACUACAUCGUGUGCAACAUCUCAAAUAUUAAAGUGUAUACA--------UACAGACGCUAG--UGUCAAGCAGCCUAAAUGCACAAAUCUCGAGACAUAAAUCACAAUUUCU ................(((.......((((..........((((....)--------))).((((....--))))..(((.......))).........)))).......)))....... ( -14.94) >DroYak_CAF1 68878 120 - 1 CCAAGGAAACUAAGUCGUGUGAAUCAUCUCAAAUAUUAAAGUGUAUACAUCCAUAUAUACAUACGCUUGUGUGUCAAGCAGCCUAAAUGUACAAAUCUUGAGACAUAAAUCACAAUUUCU ...((....))......(((((....((((((........(((((((......)))))))....(((((.....)))))..................))))))......)))))...... ( -23.80) >consensus ACAAGGAAACUAAAUCGUGUGCAUCAUCUCAAAU___AAAGUGUAUA______UAUAUACAUACGCUUG__AGUCAAGCAGCCUAAAUGUACAAAUCUCGAGACAUAAAUCACAAUUUCU .........((......(((((((................(((((((........)))))))..(((((.....))))).......))))))).......)).................. (-12.82 = -14.06 + 1.24)



| Location | 12,947,923 – 12,948,030 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.45 |
| Mean single sequence MFE | -20.26 |
| Consensus MFE | -14.92 |
| Energy contribution | -16.32 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.74 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.508698 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12947923 107 - 23771897 --ACUUCUUCAAAGUUUGCUUGCCAAACUACUUAGGUUCAACAAGGAAACUAAAUCGUGUGCAUCAUCUCAAAU---AUAUUGUAUA------UAUAUACAUACGCUUG--AGUCAAGCA --..........(((((.((((...((((.....))))...)))).))))).....(((((((...........---....))))))------)..........(((((--...))))). ( -18.96) >DroSec_CAF1 65150 96 - 1 --ACUUCUUCAAAGUUUGCUUGCCAAACUACUUAGGUUCAACAAGGAAACUAAAUCGUGUGCAUU------------AAAGUGUAUA--------UAUACAUACGCUUG--AGUCAAGCA --..........(((((.((((...((((.....))))...)))).))))).....((((((((.------------...)))))))--------)........(((((--...))))). ( -21.20) >DroSim_CAF1 69502 98 - 1 --ACUUCUUCAAAGUUUGCUUGCCAAACUACUUAGGUUCAACAAGGAAACUAAAUCGUGUGCAUU------------AAAGUGUAUA------UAUAUACAUUCGCUUG--AGUCAAGCA --..........(((((.((((...((((.....))))...)))).))))).....((((((((.------------...)))))))------)..........(((((--...))))). ( -21.20) >DroEre_CAF1 71197 110 - 1 UAACUCCUUCAAAGUUUGCUUGCCAAACUACUUAGGUUCACCAUCAAAACUACAUCGUGUGCAACAUCUCAAAUAUUAAAGUGUAUACA--------UACAGACGCUAG--UGUCAAGCA ................((((((.(..........((....))......(((((.(((((((..((((.............))))...))--------))).)).).)))--)).)))))) ( -15.72) >DroYak_CAF1 68918 120 - 1 UAACUUCUUCAAAGUUUGCUUGCCAAACUACUUAGGUUCACCAAGGAAACUAAGUCGUGUGAAUCAUCUCAAAUAUUAAAGUGUAUACAUCCAUAUAUACAUACGCUUGUGUGUCAAGCA ............((((((.....)))))).....((((((((.((....)).....).)))))))...............(((((((......)))))))....(((((.....))))). ( -24.20) >consensus __ACUUCUUCAAAGUUUGCUUGCCAAACUACUUAGGUUCAACAAGGAAACUAAAUCGUGUGCAUCAUCUCAAAU___AAAGUGUAUA______UAUAUACAUACGCUUG__AGUCAAGCA ............(((((.((((...((((.....))))...)))).))))).............................(((((((........)))))))..(((((.....))))). (-14.92 = -16.32 + 1.40)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:22:58 2006