
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,946,652 – 12,946,812 |
| Length | 160 |
| Max. P | 0.984076 |

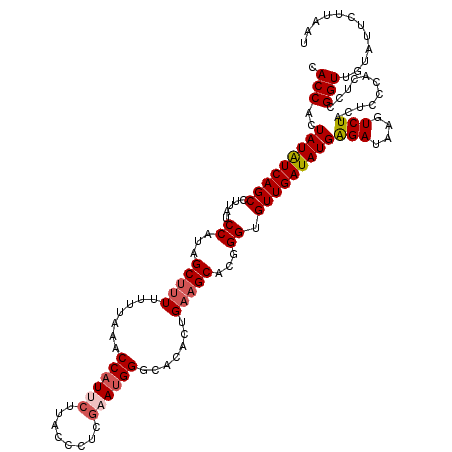
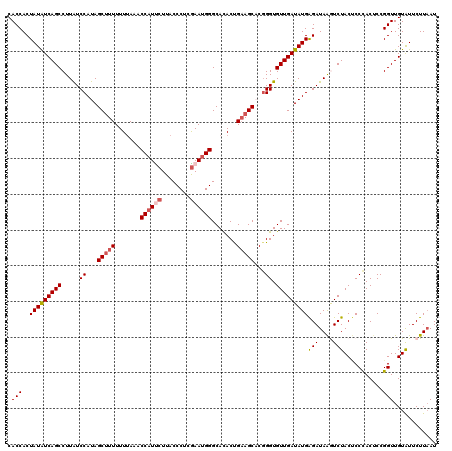
| Location | 12,946,652 – 12,946,772 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.92 |
| Mean single sequence MFE | -27.52 |
| Consensus MFE | -20.86 |
| Energy contribution | -21.66 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.782884 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12946652 120 - 23771897 CACCACUAUAUCAGCCUUAUCCAUAGCUUUUUUUAAACCAUUCUUACCCUCGAAUGGGCACACUGAAGCACGGGUGUUGAUAUGAGAUAAGUCUACUCCUACACCGGUUGUAUUCUUAAU ......(((((((((...((((...(((((.......((((((........)))))).......)))))..)))))))))))))...((((..(((.((......))..)))..)))).. ( -29.94) >DroSec_CAF1 63908 120 - 1 CACCACUAUAUCAGCCUUAUCCAUAGCUUUUUUUAAACCAUUCUUACCCUCAAAUGGGCACACUGAAGCCCGGGUGUUGAUAUGAGAUAAGUCUACUCCCACUCCGGUUGUAUUCUUAAU ......(((((((((...((((...(((((.......(((((..........))))).......)))))..)))))))))))))...((((..(((..((.....))..)))..)))).. ( -26.44) >DroSim_CAF1 68260 120 - 1 CACCACUAUAUCAGCCUUAUCCAUAGCUUUUUUUAAACCAUUCUUACCCUCGAAUGGGCAUACUGAAGCCCUGGUGUUGAUAUGAGAUAAGUCUACUCCCACUCCGGUUGUAUUCUUAAU ......(((((((((.....(((..(((((.......((((((........)))))).......)))))..))).)))))))))...((((..(((..((.....))..)))..)))).. ( -31.24) >DroEre_CAF1 69974 120 - 1 CACCACUAUAUCAGCCUUGUCCAUAGCCUUUUUUAAACCAUCCUUACCCUCGAAUGGGCACACCGAAGCACGGGUGUUGAUAUGGGAUAGAUCUACCCCUACUCCGGUUGUGUUCUUUAU (((.((((((((((((((((.................((((.(........).))))((........))))))).))))))))((..(((........)))..))))).)))........ ( -25.70) >DroYak_CAF1 67632 120 - 1 CACCACUAUGUCAGCCUUAUCCAUAGCUUUUUUCAAACCAUCCUUACCCUCGAACGGGCACACCGUAGCACGGGUGUUGAUAUGGGAUAGAUCUACGCCCACUCCGGUUGUAUUCUUAAU .....((((((((((((.......))...................((((....((((.....)))).....))))))))))))))...(((..((((.((.....)).)))).))).... ( -24.30) >consensus CACCACUAUAUCAGCCUUAUCCAUAGCUUUUUUUAAACCAUUCUUACCCUCGAAUGGGCACACUGAAGCACGGGUGUUGAUAUGAGAUAAGUCUACUCCCACUCCGGUUGUAUUCUUAAU .(((..(((((((((.....((...(((((.......((((((........)))))).......)))))...)).)))))))))(((....)))...........)))............ (-20.86 = -21.66 + 0.80)

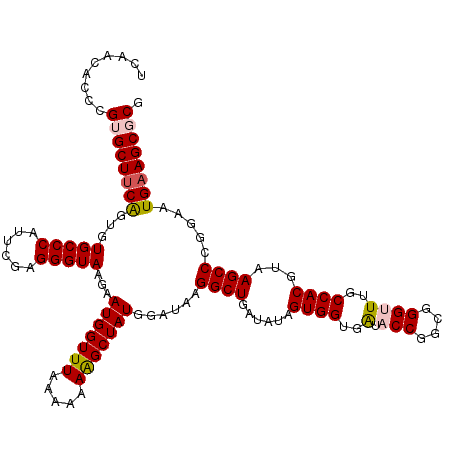
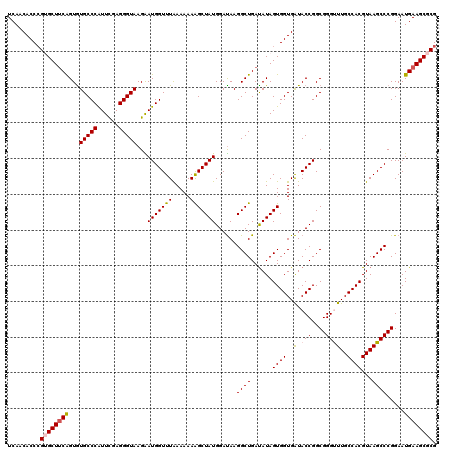
| Location | 12,946,692 – 12,946,812 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.67 |
| Mean single sequence MFE | -41.72 |
| Consensus MFE | -36.30 |
| Energy contribution | -36.74 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.984076 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12946692 120 + 23771897 UCAACACCCGUGCUUCAGUGUGCCCAUUCGAGGGUAAGAAUGGUUUAAAAAAAGCUAUGGAUAAGGCUGAUAUAGUGGUGAUACCGGCGGGUUUGCCACGUAAGCCCGGAAUGAAGCGCG ........(((((((((...(((((......)))))...(((((((.....)))))))......((((......((((..(.(((....))))..))))...)))).....))))))))) ( -40.90) >DroSec_CAF1 63948 120 + 1 UCAACACCCGGGCUUCAGUGUGCCCAUUUGAGGGUAAGAAUGGUUUAAAAAAAGCUAUGGAUAAGGCUGAUAUAGUGGUGAUACCGGCGGGUUUGCCACGUAAGCCCGGAAUGAAGCGCG .......((((((((((((.(((((......)))))...(((((((.....))))))).......)))))....((((..(.(((....))))..))))...)))))))........... ( -42.20) >DroSim_CAF1 68300 120 + 1 UCAACACCAGGGCUUCAGUAUGCCCAUUCGAGGGUAAGAAUGGUUUAAAAAAAGCUAUGGAUAAGGCUGAUAUAGUGGUGAUACCGGCGGGUUUGCCACGUAAGCCCGGAAUGAAGCGCG (((...((.((((((((((.(((((......)))))...(((((((.....))))))).......)))))....((((..(.(((....))))..))))...)))))))..)))...... ( -38.90) >DroEre_CAF1 70014 120 + 1 UCAACACCCGUGCUUCGGUGUGCCCAUUCGAGGGUAAGGAUGGUUUAAAAAAGGCUAUGGACAAGGCUGAUAUAGUGGUGGUACCGGCGGGCUUGCCACGUAAGCCCGGAAUGAAGCGCG ........(((((((((((((((((......)))))...(((((((.....)))))))...((..((((...))))..)).))))..(((((((((...)))))))))....)))))))) ( -43.40) >DroYak_CAF1 67672 120 + 1 UCAACACCCGUGCUACGGUGUGCCCGUUCGAGGGUAAGGAUGGUUUGAAAAAAGCUAUGGAUAAGGCUGACAUAGUGGUGAUACCGGCGGGCUUGCCACGUAAGCCCGGAAUGAAGCGCG ........((((((.((((((((((((((........)))))...........((((((...........))))))))).)))))).(((((((((...)))))))))......)))))) ( -43.20) >consensus UCAACACCCGUGCUUCAGUGUGCCCAUUCGAGGGUAAGAAUGGUUUAAAAAAAGCUAUGGAUAAGGCUGAUAUAGUGGUGAUACCGGCGGGUUUGCCACGUAAGCCCGGAAUGAAGCGCG .........((((((((...(((((......)))))...(((((((.....)))))))......((((......((((..(.(((....))))..))))...)))).....)))))))). (-36.30 = -36.74 + 0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:22:51 2006