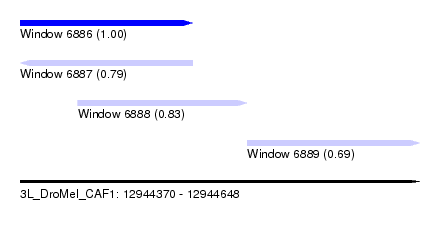
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,944,370 – 12,944,648 |
| Length | 278 |
| Max. P | 0.997251 |

| Location | 12,944,370 – 12,944,490 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.50 |
| Mean single sequence MFE | -37.12 |
| Consensus MFE | -34.94 |
| Energy contribution | -34.94 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.82 |
| SVM RNA-class probability | 0.997251 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12944370 120 + 23771897 GCCACUUAACUGAGACACCUUUUUGGGGGCCAUGUUUUGUUAUUUGGAAAUACAUUUUCUGGCCAACAUGUUGCCCGAAAUGCCUGGGCUUAAUUUAUAUGUUGUCAGACCGGGUGGCAG (((((((..((((....(((....)))(((((.....(((...........))).....)))))(((((((.(((((.......))))).......))))))).))))...))))))).. ( -36.50) >DroSec_CAF1 61621 120 + 1 GCCACUUAACUGAGACACCUUUUUGGGGCCCAUGUUUUGUUAUUUGGAAAUACAUUUUCUGGCCAACAUGUUGCCCGAAAUGCCUGGGCUUAAUUUAUAUGUUGUCAGACCGGGUGGCAG (((((((..((((((((((.....))(((((((((((((......(((((.....)))))(((.((....))))))))))))..)))))).........)))).))))...))))))).. ( -35.60) >DroSim_CAF1 65967 120 + 1 GCCACUUAACUGAGACACCUUUUUGGGGCCCAUGUUUUGUUAUUUGGAAAUACAUUUUCUGGCCAACAUGUUGCCCGAAAUGCCUGGGCUUAAUUUAUAUGUUGUCAGACCGGGUGGCAG (((((((..((((((((((.....))(((((((((((((......(((((.....)))))(((.((....))))))))))))..)))))).........)))).))))...))))))).. ( -35.60) >DroEre_CAF1 67850 120 + 1 GCCACUUAACUGAGACACCUUCUUGGGAGCCGUGUUUUGUUAUUUGGAAAUACAUUCUCUGGACAACAUGUUGCCCGAAAUGCCUGGGCUUUAUUUAUAUGUUGUCAGACUGGGUGGCAG ((((((((.(.((((..(((....)))....(((((((........)))))))..)))).)((((((((((.(((((.......))))).......))))))))))....)))))))).. ( -41.10) >DroYak_CAF1 65436 120 + 1 GCCACUUAACUGAGACACCUUUUUGGGACCCAUGUUUUGUUUUUUGGAAAUACAUUUUCUGGACAACAUGUUGCCCGAAAUGCCUGGGCUUAAUUUAUAUGUUGUCAGACCGGGUGGCAG (((((((..((((((((...(((((((...(((((..(((((...(((((.....)))))))))))))))...))))))).((....))..........)))).))))...))))))).. ( -36.80) >consensus GCCACUUAACUGAGACACCUUUUUGGGGCCCAUGUUUUGUUAUUUGGAAAUACAUUUUCUGGCCAACAUGUUGCCCGAAAUGCCUGGGCUUAAUUUAUAUGUUGUCAGACCGGGUGGCAG ((((((((((.(((((((((....))).....)))))))))................(((((.((((((((.(((((.......))))).......)))))))))))))..))))))).. (-34.94 = -34.94 + 0.00)

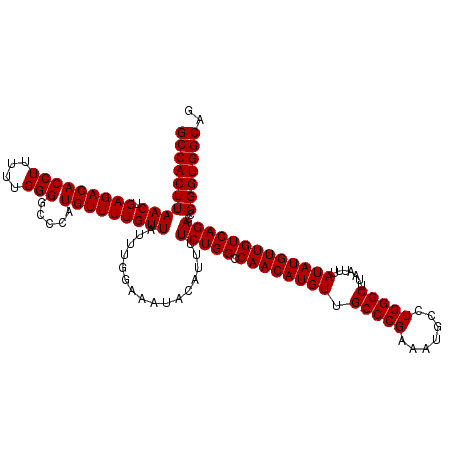

| Location | 12,944,370 – 12,944,490 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.50 |
| Mean single sequence MFE | -30.11 |
| Consensus MFE | -25.54 |
| Energy contribution | -25.74 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.785201 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12944370 120 - 23771897 CUGCCACCCGGUCUGACAACAUAUAAAUUAAGCCCAGGCAUUUCGGGCAACAUGUUGGCCAGAAAAUGUAUUUCCAAAUAACAAAACAUGGCCCCCAAAAAGGUGUCUCAGUUAAGUGGC ..(((((.....((((..((((.........((((.........))))....((..(((((.....((((((....))).))).....)))))..)).....)))).))))....))))) ( -29.20) >DroSec_CAF1 61621 120 - 1 CUGCCACCCGGUCUGACAACAUAUAAAUUAAGCCCAGGCAUUUCGGGCAACAUGUUGGCCAGAAAAUGUAUUUCCAAAUAACAAAACAUGGGCCCCAAAAAGGUGUCUCAGUUAAGUGGC ..(((((.....((((..((((.........(((((.(((((((((.(((....))).)).).))))))...................))))).........)))).))))....))))) ( -30.32) >DroSim_CAF1 65967 120 - 1 CUGCCACCCGGUCUGACAACAUAUAAAUUAAGCCCAGGCAUUUCGGGCAACAUGUUGGCCAGAAAAUGUAUUUCCAAAUAACAAAACAUGGGCCCCAAAAAGGUGUCUCAGUUAAGUGGC ..(((((.....((((..((((.........(((((.(((((((((.(((....))).)).).))))))...................))))).........)))).))))....))))) ( -30.32) >DroEre_CAF1 67850 120 - 1 CUGCCACCCAGUCUGACAACAUAUAAAUAAAGCCCAGGCAUUUCGGGCAACAUGUUGUCCAGAGAAUGUAUUUCCAAAUAACAAAACACGGCUCCCAAGAAGGUGUCUCAGUUAAGUGGC ..(((((.....((((..((((.........((....)).((((((((((....)))))).))))))))....................(((.((......)).)))))))....))))) ( -29.70) >DroYak_CAF1 65436 120 - 1 CUGCCACCCGGUCUGACAACAUAUAAAUUAAGCCCAGGCAUUUCGGGCAACAUGUUGUCCAGAAAAUGUAUUUCCAAAAAACAAAACAUGGGUCCCAAAAAGGUGUCUCAGUUAAGUGGC ..(((((..((....(((.............((....)).((((((((((....)))))).)))).)))....)).........(((.((((.(((.....)).).)))))))..))))) ( -31.00) >consensus CUGCCACCCGGUCUGACAACAUAUAAAUUAAGCCCAGGCAUUUCGGGCAACAUGUUGGCCAGAAAAUGUAUUUCCAAAUAACAAAACAUGGGCCCCAAAAAGGUGUCUCAGUUAAGUGGC ..(((((.....((((..((((.........(((((.(((((((((.(((....))).)).))).))))...................))))).........)))).))))....))))) (-25.54 = -25.74 + 0.20)


| Location | 12,944,410 – 12,944,528 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.41 |
| Mean single sequence MFE | -32.92 |
| Consensus MFE | -26.92 |
| Energy contribution | -27.12 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.830053 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12944410 118 + 23771897 UAUUUGGAAAUACAUUUUCUGGCCAACAUGUUGCCCGAAAUGCCUGGGCUUAAUUUAUAUGUUGUCAGACCGGGUGGCAGCAACAAAUGCAGC--CGCAAUAGUGGAAUUACUCAUACAC ............((((.((((((.(((((((.(((((.......))))).......)))))))))))))....(((((.(((.....))).))--)))...))))............... ( -31.70) >DroSec_CAF1 61661 116 + 1 UAUUUGGAAAUACAUUUUCUGGCCAACAUGUUGCCCGAAAUGCCUGGGCUUAAUUUAUAUGUUGUCAGACCGGGUGGCAGCAACAAAUGCAGC--CGCAAUAGUGGAAUUACUCA--CAU ...((((..........((((((.(((((((.(((((.......))))).......)))))))))))))))))(((((.(((.....))).))--)))....((((......)))--).. ( -33.50) >DroSim_CAF1 66007 116 + 1 UAUUUGGAAAUACAUUUUCUGGCCAACAUGUUGCCCGAAAUGCCUGGGCUUAAUUUAUAUGUUGUCAGACCGGGUGGCAGCAACAAAUGCAGC--CGCAAUAGUGGAAUUACUCA--CAC ...((((..........((((((.(((((((.(((((.......))))).......)))))))))))))))))(((((.(((.....))).))--)))....((((......)))--).. ( -33.50) >DroEre_CAF1 67890 114 + 1 UAUUUGGAAAUACAUUCUCUGGACAACAUGUUGCCCGAAAUGCCUGGGCUUUAUUUAUAUGUUGUCAGACUGGGUGGCAGCAACAAAUGCAGC--CGCAAUAGUGGAAUUACUCA--C-- .............(((((...((((((((((.(((((.......))))).......))))))))))..((((.(((((.(((.....))).))--)))..)))))))))......--.-- ( -33.20) >DroYak_CAF1 65476 118 + 1 UUUUUGGAAAUACAUUUUCUGGACAACAUGUUGCCCGAAAUGCCUGGGCUUAAUUUAUAUGUUGUCAGACCGGGUGGCAGCAACAAAUGCAGCACGGCAAUAGUGGAAUUACUCA--CAU .....(((((.....))))).((((((((((.(((((.......))))).......)))))))))).(.(((.((.(((........))).)).))))....((((......)))--).. ( -32.70) >consensus UAUUUGGAAAUACAUUUUCUGGCCAACAUGUUGCCCGAAAUGCCUGGGCUUAAUUUAUAUGUUGUCAGACCGGGUGGCAGCAACAAAUGCAGC__CGCAAUAGUGGAAUUACUCA__CAC .................(((((.((((((((.(((((.......))))).......)))))))))))))..(((((((.(((.....))).))..(((....)))....)))))...... (-26.92 = -27.12 + 0.20)

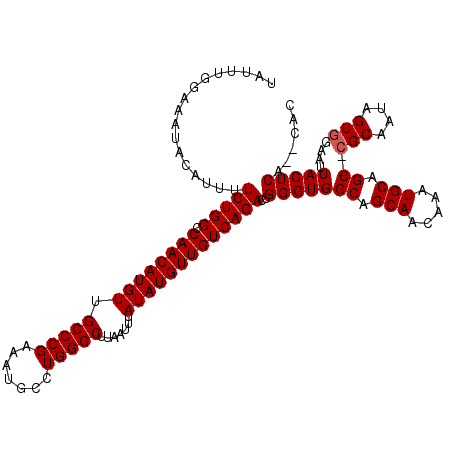

| Location | 12,944,528 – 12,944,648 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.78 |
| Mean single sequence MFE | -37.70 |
| Consensus MFE | -31.52 |
| Energy contribution | -31.32 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.693451 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12944528 120 + 23771897 ACACACGCUCAAUCUGAAACGUGGCCAGGUGAACUCCUGGCGAAAGUGAAACUGGGUCGCAAGAAAGUUGUCGGGCCACCACCCACUUUCCUUUUUGGCAAUGGGGGGGACAACAAGAUA ...((((.((.....))..))))((((((......))))))....((((.......))))......(((((((((......))).((..((...........))..))))))))...... ( -37.80) >DroSec_CAF1 61777 119 + 1 ACACACGCUCAAUUCGAAACUUGGCCAGGUGAACUCCUGGCGAAAGUGAAACUGGGUCGCAAGAAAGUUGUCGGGCCACCACCCACUUUCCUUUUGGGCAAUGGG-GGGACAACAAGAUA .......((((.((((....((.((((((......)))))).))..))))..))))((....))..((((((((....)).((((.(.(((....))).).))))-..))))))...... ( -38.10) >DroSim_CAF1 66123 119 + 1 ACACACGCUCCAUCUGAAACUUGGCCAGGUGAACUCCUGGCGAAAGUGAAACUGGGUCGCAAGAAAGUUGUCGGGCCACCACCCACUUUCCUUUUGGGCAAUGGG-GGGACAACAAGAUA ........((((......((((.((((((......))))))..)))).....))))((....))..((((((((....)).((((.(.(((....))).).))))-..))))))...... ( -39.30) >DroEre_CAF1 68004 114 + 1 --ACACGCUCAAUCUGAAACUUGGCCAGGUGAACUCCUGGCGAAAGUGAAACUGGGUCGCAAGAAAGUUUUCGGGCC---ACCCACUUUCCUUUUUGGCAAUGGG-GGGACAACGAGAUA --.....(((..(((((.((((.((((((......))))))..)))).(((((...((....)).))))))))))((---.((((.((.((.....)).))))))-))......)))... ( -37.20) >DroYak_CAF1 65594 116 + 1 ACACACGCCCAAUCUGAAACUUGGCCAGGUGAACUCUUGGUGAAAGUGAAACUGGGUCGCAAGAAAGUUUUCGGGCC---ACCCACUUUCUUUUUUGGCACUGGG-GGUACAACAAGAUA ....((.((((..(..((((((.((((((......))))))..))))((((.(((((.((..(((....)))..)).---))))).))))...))..)...))))-.))........... ( -36.10) >consensus ACACACGCUCAAUCUGAAACUUGGCCAGGUGAACUCCUGGCGAAAGUGAAACUGGGUCGCAAGAAAGUUGUCGGGCCACCACCCACUUUCCUUUUUGGCAAUGGG_GGGACAACAAGAUA .....(.((((..((((((....((((((......))))))((((((((((((...((....)).)))).))(((......)))))))))..))))))...)))).)............. (-31.52 = -31.32 + -0.20)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:22:46 2006